- published: 31 Dec 2013
- views: 167781
-
remove the playlistBiochemistry
- remove the playlistBiochemistry
- published: 06 Jan 2015
- views: 85500
- published: 05 Dec 2012
- views: 102562
- published: 13 Feb 2012
- views: 1733264
- published: 20 Mar 2013
- views: 33799
- published: 25 Jan 2015
- views: 5234
- published: 07 Oct 2013
- views: 33909
- published: 03 Apr 2013
- views: 22785
- published: 14 Apr 2013
- views: 20679
- published: 14 Jul 2015
- views: 2766
- published: 01 Nov 2014
- views: 8023

Biochemistry, sometimes called biological chemistry, is the study of chemical processes in living organisms, including, but not limited to, living matter. Biochemistry governs all living organisms and living processes. By controlling information flow through biochemical signalling and the flow of chemical energy through metabolism, biochemical processes give rise to the complexity of life.
Much of biochemistry deals with the structures, functions and interactions of cellular components such as proteins, carbohydrates, lipids, nucleic acids and other biomolecules —although increasingly processes rather than individual molecules are the main focus. Among the vast number of different biomolecules, many are complex and large molecules (called biopolymers), which are composed of similar repeating subunits (called monomers). Each class of polymeric biomolecule has a different set of subunit types. For example, a protein is a polymer whose subunits are selected from a set of 20 or more amino acids. Biochemistry studies the chemical properties of important biological molecules, like proteins, and in particular the chemistry of enzyme-catalyzed reactions.
This article is licensed under the Creative Commons Attribution-ShareAlike 3.0 Unported License, which means that you can copy and modify it as long as the entire work (including additions) remains under this license.
- Loading...

-
 3:49
3:49Introduction to Biochemistry HD
Introduction to Biochemistry HDIntroduction to Biochemistry HD
This is a new high definition (HD) dramatic video choreographed to powerful music that introduces the viewer/student to Biochemistry. It is designed as a motivational "trailer" to be shown by teachers in Biology, Biochemistry and Chemistry classrooms in middle school, high school and college as a visual Introduction to the wonders of the Biochemistry of life. It replaces an earlier video on the same topic that I produced over 3 years ago. Subscribe to my channel at http://www.youtube.com/user/sfgregs?feature=mhum to see all of my exciting video trailers in Biology, Chemistry, Earth Science and Astronomy. I will be releasing new ones periodically. You can download all of my videos for free from Vimeo, my other video site. The link is available in the "About" section of this channel. Please rate this video and feel free to comment. If you like it, please help me spread the word by posting links to it on your school and social media websites. The more students who can enjoy these dramatic videos, the better! I wish to thank all the quality video and music producers whose postings enabled me to assemble this video for educational use. To best enjoy this video, view on a big screen and turn up your speakers. The music is powerful and dramatic! I can customize this video to add your name or school name at the end credits, for a very modest fee. If interested, email me at "fsgregs@comcast.net". -
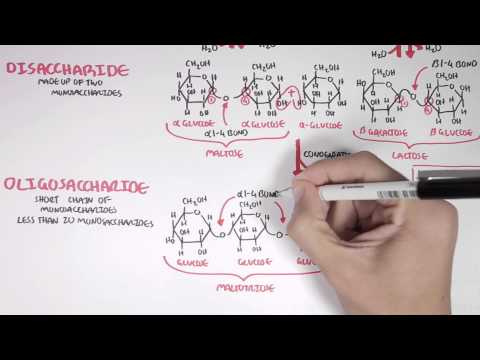 16:15
16:15Biochemistry of Carbohydrates
Biochemistry of CarbohydratesBiochemistry of Carbohydrates
Video was part of 2014 Summer Scholarship Project with CSIRO called "The Hungry Microbiome" For more visit: http://www.csiro.au/hungrymicrobiome/ https://www.facebook.com/ArmandoHasudungan Support me: http://www.patreon.com/armando Instagram: http://instagram.com/armandohasudungan Twitter: https://twitter.com/Armando71021105 -
 9:12
9:12How to Study Biochemistry in Medical School
How to Study Biochemistry in Medical SchoolHow to Study Biochemistry in Medical School
Link to the books in this video on Amazon: Rapid Review Biochemistry: http://www.amazon.com/gp/product/0323068871/ref=as_li_ss_tl?ie=UTF8&camp;=1789&creative;=390957&creativeASIN;=0323068871&linkCode;=as2&tag;=doctheundfor-20 Lippincott's Biochemistry http://www.amazon.com/gp/product/160831412X/ref=as_li_ss_tl?ie=UTF8&camp;=1789&creative;=390957&creativeASIN;=160831412X&linkCode;=as2&tag;=doctheundfor-20 Lehninger Principles of Biochemistry http://www.amazon.com/gp/product/071677108X/ref=as_li_ss_tl?ie=UTF8&camp;=1789&creative;=390957&creativeASIN;=071677108X&linkCode;=as2&tag;=doctheundfor-20 -
 14:09
14:09Biological Molecules - You Are What You Eat: Crash Course Biology #3
Biological Molecules - You Are What You Eat: Crash Course Biology #3Biological Molecules - You Are What You Eat: Crash Course Biology #3
Hank talks about the molecules that make up every living thing - carbohydrates, lipids, and proteins - and how we find them in our environment and in the food that we eat. Crash Course Biology is now available on DVD! http://dftba.com/product/1av/CrashCourse-Biology-The-Complete-Series-DVD-Set Follow CrashCourse on Twitter: http://www.twitter.com/thecrashcourse Like CrashCourse on Facebook: http://www.facebook.com/youtubecrashcourse Resources for this episode in the Google Document here: http://dft.ba/-citations2 TAGS: biological molecules, carbohydrates, lipids, proteins, nucleic acids, food, biolography, william prout, urea, energy, monosaccharides, glucose, fructose, disaccharides, sucrose, polysaccharides, simple sugars, cellulose, starch, glycogen, glycerol, fatty acid, triglyceride, phospholipid, steroid, cholesterol, enzymes, antibodies, hormones, amino acids, nitrogen, polypeptides, protein synthesis, biology, molecule, crashcourse, hank green Support CrashCourse on Subbable: http://subbable.com/crashcourse -
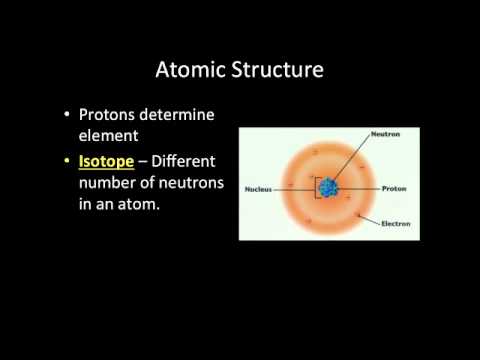 10:23
10:23Video Lecture: Introduction to Biochemistry
Video Lecture: Introduction to BiochemistryVideo Lecture: Introduction to Biochemistry
Introductory topics related to biochemistry. Atoms, molecules, and the basis of chemistry is discussed by Mr. Joseph Lamb -
 15:29
15:29Biochemistry 4.0: Introduction to amino acids
Biochemistry 4.0: Introduction to amino acidsBiochemistry 4.0: Introduction to amino acids
Includes a review of protein function, the basics of stereochemistry (L- and D-amino acids) and a run through of all amino acids found in proteins. -
 12:24
12:24Biochemistry (Molecular and Cellular) at Oxford University
Biochemistry (Molecular and Cellular) at Oxford UniversityBiochemistry (Molecular and Cellular) at Oxford University
Want to know more about studying at Oxford University? Watch this short film to hear tutors and students talk about this undergraduate degree. For more information on this course, please visit our website at http://www.ox.ac.uk/admissions/undergraduate_courses/courses/biochemistry_molecular_and_cellular/biochemistry.html -
 49:33
49:33#1 Biochemistry Lecture (Introduction) from Kevin Ahern's BB 350
#1 Biochemistry Lecture (Introduction) from Kevin Ahern's BB 350#1 Biochemistry Lecture (Introduction) from Kevin Ahern's BB 350
1. Contact me at kgahern@davincipress.com / Friend me on Facebook (kevin.g.ahern) 2. Download my free biochemistry book at http://biochem.science.oregonstate.edu/biochemistry-free-and-easy 3. Take my free iTunes U course at https://itunes.apple.com/us/course/biochemistry/id556410409 4. Check out my free book for pre-meds at http://biochem.science.oregonstate.edu/biochemistry-free-and-easy 5. Lecturio videos for medical students - https://www.lecturio.com/medical-courses/biochemistry.course 6. Course video channel at http://www.youtube.com/user/oharow/videos?view=1 7. Check out all of my free workshops at http://oregonstate.edu/dept/biochem/ahern/123.html 8. Check out my Metabolic Melodies at http://www.davincipress.com/ 9. My courses can be taken for credit (wherever you live) via OSU's ecampus. For details, see http://ecampus.oregonstate.edu/soc/ecatalog/ecourselist.htm?termcode=all&subject;=BB 10. Course materials at http://oregonstate.edu/instruct/bb350 Lecture Highlights 1. All students need to read the course syllabus. You can download it from the Schedule page. 2. Biochemistry as a science is relatively new, at least in its present form. 3. Our understanding of the basis of life has evolved from examining the organism as the foundation of life down to cells, then down to organelles, and then finally down to molecules. 4. Molecules are the foundation of life and are the basis of molecular biology. 5. Prokaryotes (bacteria) differ from eukaryotes in the composition of their cells. Eukyaryotes have organelles and prokaryotes do not. Eukaryotes include yeast (single celled, dog, cats, plants, humans, and more). All prokaryotes are unicellular. 6. The nucleus holds DNA, chromosomes, and RNA. The mitochondrion is involved in energy production. The chloroplast is involved in photosynthesis in plants. 7. Cells have a cytoskeleton, which to a cell what a skeleton is to you. 8. Cells require energy to exist. There are many reasons for this, but one is the need to counter the forces of entropy. 9. Cells store energy in ATP for immediate use. The potential energy stored in ATP arises from the replusion of the negatively charged phosphates for each other. 10. The building blocks of DNA and RNA are nucleotides. The building blocks of protein are amino acids. There are 20 amino acids in proteins. -
 9:18
9:18A day in the life of a biochemistry student
A day in the life of a biochemistry studentA day in the life of a biochemistry student
Ever wonder what the life of a science major, particularly a biochemistry student, is really like? -Biochemistry, B.S. (May 2013) California State University, Fullerton Music: Walking on Sunshine - Katrina and the Waves Carmina Burana: O Fortuna - Carl Orff Also Sprach Zarathustr - Richard Strauss Rawhide Remix - Blues Brothers Cats on Mars - Cowboy Bepop Time (Inception) - Hans Zimmer **the last scene is directly related to Inception** i.e. dreams within dreams of trying to solve puzzles representing finishing that o chem hw in real life -
 4:34
4:34CAREERS IN B.SC BIOCHEMISTRY– M.Sc,P.Hd,Research Institutes,Job Opportunities,Salary Package
CAREERS IN B.SC BIOCHEMISTRY– M.Sc,P.Hd,Research Institutes,Job Opportunities,Salary PackageCAREERS IN B.SC BIOCHEMISTRY– M.Sc,P.Hd,Research Institutes,Job Opportunities,Salary Package
CAREERS IN B.SC BIOCHEMISTRY.Go through the career opportunities of B.SC BIOCHEMISTRY, Govt jobs and Employment News channel from Freshersworld.com – The No.1 job portal for freshers in India. Visit http://www.freshersworld.com?src=Youtube for detailed Career information,Job Opportunities,Education details of B.SC BIOCHEMISTRY. Bachelors in Science in Biochemistry is an exciting venture these days. There are quite a few job openings for this field, and there are some good courses available for further studies. We shall discuss a few career opportunities available to BSc graduates and also a few options available for higher studies. Biochemistry involves the study of chemical and biological processes in organisms at a very minute level. The course offers you insights into various biological and chemical processes that takes place at a cellular level, you will also have a brief idea about the effect of environment on organisms as well. You can find employment opportunities at both the public and private sectors in India. Let us first discuss the various private sector jobs you can find in this domain. The scope of work for BSc graduate in biochemistry is wide, and you can find yourself a job in laboratories, pharmaceutical companies, and even in hospitals. There are quite a few job descriptions you can apply to. Some of them are Geneticist, anaesthesiologist, Chemist, Cytologist, Clinical research specialist, Patent attorney, laboratory supervisor, regulatory affairs specialist, toxicologist and perfumer. You can find employment at many companies too including: 1. Agricultural Research Services 2. Biotechnology Firms 3. Chemical Industry 4. Chemicals Manufacturing Companies 5. Colleges / Universities 6. Cosmetic Companies 7. Education Sector 8. Engineering Firms 9. Food Institutes 10. Forensic Crime Research 11. Health Care Providers 12. Heavy Chemical Industries 13. Hospitals 14. Indian Civil Services 15. Industrial Laboratories 16. Laboratories 17. Manufacturing and Processing Firms 18. Medical Laboratories 19. Medical Research 20. Oil Industry 21. Petroleum Companies 22. Pharmaceutical Companies 23. Power Generating Companies 24. Pyrotechnics Manufacturers 25. Research and Development Firms 26. Schools 27. Seed And Nursery Companies 28. Technical Journals 29. Testing Laboratories 30. Utility Companies 31. Waste-water Plants You can find many job opportunities in the public sector too. You can find jobs through UPSC, State level PSC’s and SSCs. You can use your knowledge effectively at various government institution, primarily those which are related heavily with biology and chemistry. Some of the government institutions where you can find jobs are 1. Agriculture and fisheries 2. Blood Service 3. Cancer research institutes 4. Environmental Pollution Control 5. Forensic Science 6. Hospitals 7. National Blood Services 8. Overseas Development 9. Public Health Entities 10. Public Health Laboratories You can even take up a teaching position before you decide about your future. The incentives are very good, so long as a teaching career is concerned and the pay scale these days for teachers is really good too. You can even prefer to continue your education and pursue a degree in MSc in the field of biochemistry from many reputed colleges in India or abroad. It was observed that there are more employment opportunities for people who have finished their master’s degree as compared to those with only a bachelor’s degree in Biochemistry. For more jobs & career information and daily job alerts, subscribe to our channel and support us. You can also install our Mobile app for govt jobs for getting regular notifications on your mobile. Freshersworld.com is the No.1 job portal for freshers jobs in India. Check Out website for more Jobs & Careers. http://www.freshersworld.com?src=Youtube - - ***Disclaimer: This is just a career guidance video for fresher candidates. The name, logo and properties mentioned in the video are proprietary property of the respective companies. The career and job information mentioned are an indicative generalised information. In no way Freshersworld.com, indulges into direct or indirect recruitment process of the respective companies. -
 15:02
15:02Biochemistry Basics
Biochemistry BasicsBiochemistry Basics
Biochemistry Basics -
 5:20
5:20What is biochemistry?
What is biochemistry?What is biochemistry?
A brief and concise introduction to biochemistry. What is it? What does it involve? And why is it important? -
 14:47
14:47Introduction to Biochemistry Lecture by Kevin Ahern, Part 1 of 4
Introduction to Biochemistry Lecture by Kevin Ahern, Part 1 of 4Introduction to Biochemistry Lecture by Kevin Ahern, Part 1 of 4
1. Contact me at kgahern@davincipress.com / Friend me on Facebook (kevin.g.ahern) 2. Download my free biochemistry book at http://biochem.science.oregonstate.edu/biochemistry-free-and-easy 3. Take my free iTunes U course at https://itunes.apple.com/us/course/biochemistry/id556410409 4. Check out my free book for pre-meds at http://biochem.science.oregonstate.edu/biochemistry-free-and-easy 5. Lecturio videos for medical students - https://www.lecturio.com/medical-courses/biochemistry.course 6. Course video channel at http://www.youtube.com/user/oharow/videos?view=1 7. Check out all of my free workshops at http://oregonstate.edu/dept/biochem/ahern/123.html 8. Check out my Metabolic Melodies at http://www.davincipress.com/ 9. My courses can be taken for credit (wherever you live) via OSU's ecampus. For details, see http://ecampus.oregonstate.edu/soc/ecatalog/ecourselist.htm?termcode=all&subject;=BB 10. Course materials at http://oregonstate.edu/instruct/bb350 Contact me at kgahern@davincipress.com - My books are below. FREE BIOCHEMISTRY BOOK - http://biochem.science.oregonstate.edu/biochemistry-free-and-easy HOW-TO BOOK FOR PRE-MEDS - "Kevin and Indira's Guide to Getting Into Medical School" - http://www.lulu.com/shop/product-20639977.html My Course Videos - http://www.youtube.com/user/oharow/videos?view=1 My Workshops - http://www.youtube.com/playlist?list=PLlnFrNM93wqyTiCLZKehU1Tp8rNmnOWYB&feature;=view_all My Metabolic Melodies - http://www.davincipress.com/metabmelodies.html TAKE MY COURSES for credit (wherever you live) via OSU's ecampus. For details, see http://ecampus.oregonstate.edu/soc/ecatalog/ecourselist.htm?termcode=all&subject;=BB My free iTunes courses for your iPad or iPhone Biochemistry for Pre-Meds - https://itunesu.itunes.apple.com/audit/COFY6WAD9C Elementary Biochemistry - https://itunesu.itunes.apple.com/enroll/FZD-YLQ-T9B Kevin & Indira's Guide to Getting Into Medical School - https://itunes.apple.com/us/course/id595023397 Check out this course at http://oregonstate.edu/instruction/bb350/ Related courses include BB 450 - http://oregonstate.edu/instruction/bb450/ BB 451 - http://oregonstate.edu/instruction/bb451/ BB 100 - http://oregonstate.edu/instruction/bb100/ -
 4:03
4:03BSc Biochemistry - School of Biosciences
BSc Biochemistry - School of BiosciencesBSc Biochemistry - School of Biosciences
Charlotte - an undergraduate student in the School of Biosciences - describes her experiences of the BSc Biochemistry degree course and of her time at the University of Birmingham. http://www.birmingham.ac.uk/students/courses/undergraduate/biosciences/biochemistry.aspx
- Acetal
- Acetyl-CoA
- Actin
- Activation energy
- Adaptation
- Adenine
- Aerobic glycolysis
- Alanine
- Aldehyde
- Aldoses
- Aliphatic
- Alpha helix
- Amine
- Amino
- Amino acid
- Amino Acids
- Amino group
- Ammonia
- Ammonium
- Amphiphilic
- Amylase
- Analytical chemistry
- Anatomy
- Andrew Z. Fire
- Animal
- Anselme Payen
- Antibody
- Arginine
- Aromatic
- Asparagine
- Aspartate
- Astrobiology
- Atom
- Base (chemistry)
- Beta sheet
- Biochemist
- Biochemistry
- Biocoenosis
- Biodiversity
- Biogeography
- Bioinformatics
- Biological system
- Biology
- Biomechanics
- Biomolecule
- Biomolecules
- Bioorganic chemistry
- Biophysics
- Biopolymer
- Biosphere
- Biostatistics
- Blood plasma
- Boron
- Botany
- Bromine
- Butter
- Calcium
- Cane sugar
- Carbohydrate
- Carbohydrates
- Carbon
- Carbon chauvinism
- Carbon dioxide
- Carboxyl
- Carboxyl group
- Carboxylic acid
- Carl Cori
- Carl Neuberg
- Catalysis
- Catalyze
- Category Biology
- Cell (biology)
- Cell biology
- Cell membrane
- Cell nucleus
- Cell signalling
- Cell theory
- Cell wall
- Cellulose
- Cheese
- Chemical biology
- Chemical ecology
- Chemical elements
- Chemical equilibrium
- Chemical kinetics
- Chemical physics
- Chemical reaction
- Chemist
- Chemistry
- Chemistry education
- Cholesterol
- Chromatography
- Chronobiology
- Citric acid cycle
- Click chemistry
- Cluster chemistry
- Colin Pitchfork
- Community ecology
- Conservation biology
- Cori cycle
- Craig C. Mello
- Cysteine
- Cytosine
- Dehydration reaction
- Deoxyribose
- Diastase
- Dietary mineral
- Dipeptide
- Disaccharide
- Disaccharides
- DNA
- Double bond
- Ecological niche
- Ecology
- Ecosystem
- Ecosystem ecology
- Eduard Buchner
- Edward Tatum
- Electrochemistry
- Electron microscope
- Endocrine system
- Energy
- Enzyme
- Epidemiology
- Epigenetics
- Epistasis
- Essential amino acid
- Ethanol
- Ethanol fermentation
- Ether bond
- Evolution
- Evolutionary biology
- FADH2
- Fat
- Fatty acid
- Fatty acids
- Forensic science
- Francis Crick
- Friedrich Wöhler
- Fructose
- Fruit
- Furanose
- Galactose
- Gene
- Gene flow
- Gene therapy
- Genetic code
- Genetic drift
- Genetic information
- Genetic interactions
- Genetic material
- Genetics
- Genomics
- George Beadle
- Gerty Cori
- Ghee
- Globin
- Glossary of ecology
- Gluconeogenesis
- Glucose
- Glutamate
- Glutamine
- Glycerol
- Glycine
- Glycogen
- Glycolipid
- Glycolysis
- Green chemistry
- Guanine
- Habitat
- Heme
- Hemiacetal
- Hemoglobin
- Heterocyclic
- Histidine
- Histology
- Homeostasis
- Human
- Human biology
- Human serum albumin
- Hydrogen
- Hydrolysis
- Hydrophilic
- Hydrophobic
- Hydroxyl
- Hydroxyl group
- Immunology
- Inorganic chemistry
- Iodine
- Isoleucine
- James D. Watson
- Keto acid
- Ketone
- Ketoses
- Krebs cycle
- Lactase
- Lactase deficiency
- Lactic acid
- Lactose
- Leucine
- Life
- Life sciences
- Lipid
- Lipids
- Liposome
- List of biochemists
- List of biomolecules
- Liver
- Lysine
- Macroevolution
- Macromolecule
- Marine biology
- Materials science
- Maurice Wilkins
- Medicinal chemistry
- Medicine
- Meiosis
- Membrane transport
- Metabolic pathway
- Metabolism
- Metabolome
- Metabolomics
- Methionine
- Microbiology
- Microevolution
- Milk products
- Mitosis
- Molecular Biology
- Molecular biology
- Molecular dynamics
- Molecular medicine
- Molecule
- Molecules
- Monomer
- Monosaccharide
- Monosaccharides
- Mutant
- Mutation
- Mycology
- Myosin
- NADH
- Nanobiotechnology
- Natural selection
- Neuroscience
- Neurotransmitter
- Nitrogen
- Nobel Prize
- Nonpolar
- Nuclear chemistry
- Nucleic acid
- Nucleic Acids
- Nucleic acids
- Nucleotide
- Nucleotides
- Nutrition
- Oil
- Oligosaccharide
- Organ (anatomy)
- Organelle
- Organic chemistry
- Organism
- Osteichthyes
- Oxygen
- Paleontology
- Parasitology
- Parenteral
- Pathology
- Pentose
- Peptide
- Peptide bond
- Periodic table
- Pharmacology
- Pharmacy
- Phenotype
- Phenylalanine
- Phosphate
- Phospholipid
- Phospholipids
- Phosphorus
- Photochemistry
- Photosynthesis
- Physical chemistry
- Physiology
- Plant
- Plant physiology
- Polar molecule
- Polymer chemistry
- Polysaccharide
- Polysaccharides
- Population
- Population Ecology
- Portal Biology
- Primary structure
- Proline
- Protein
- Protein synthesis
- Proteins
- Purine
- Pyranose
- Pyrimidine
- Pyruvate
- Quantum biology
- Quantum chemistry
- Quaternary structure
- Reducing end
- Resource (biology)
- Retinoid
- Ribonucleic acid
- RNA
- RNA interference
- RNAi
- Rosalind Franklin
- Saccharose
- Second messenger
- Secondary structure
- Selenium
- Serine
- Sickle-cell disease
- Signal transduction
- Silicon
- Skeletal muscle
- Small intestine
- Small molecule
- Sodium
- Speciation
- Spectroscopy
- Sphingolipid
- Steroid
- Stoichiometry
- Structural biology
- Substituent
- Sucrose
- Sugar
- Surface chemistry
- Systematics
- Systems biology
- Taxonomy
- Template Alcohols
- Template AminoAcids
- Template Biology nav
- Template Eicosanoids
- Template Fatty acids
- Template Glycosides
- Template Lipids
- Template Metabolism
- Terpenoid
- Tertiary structure
- Thermochemistry
- Threonine
- Thymine
- Tissue (biology)
- Toxicology
- Transaminase
- Transamination
- Transfersome
- Triglycerides
- Tryptophan
- Tyrosine
- Unicellular
- Uracil
- Urea
- Urea cycle
- Valine
- Vegetable oil
- Vertebrate
- Veterinary
- Water
- Wax
- Wet chemistry
- Wikiversity
- Wild type
- WP Books
- X-ray diffraction
- Yeast
- Zoology
-

Introduction to Biochemistry HD
This is a new high definition (HD) dramatic video choreographed to powerful music that introduces the viewer/student to Biochemistry. It is designed as a motivational "trailer" to be shown by teachers in Biology, Biochemistry and Chemistry classrooms in middle school, high school and college as a visual Introduction to the wonders of the Biochemistry of life. It replaces an earlier video on the same topic that I produced over 3 years ago. Subscribe to my channel at http://www.youtube.com/user/sfgregs?feature=mhum to see all of my exciting video trailers in Biology, Chemistry, Earth Science and Astronomy. I will be releasing new ones periodically. You can download all of my videos for free from Vimeo, my other video site. The link is available in the "About" section of this channel. Plea... -

Biochemistry of Carbohydrates
Video was part of 2014 Summer Scholarship Project with CSIRO called "The Hungry Microbiome" For more visit: http://www.csiro.au/hungrymicrobiome/ https://www.facebook.com/ArmandoHasudungan Support me: http://www.patreon.com/armando Instagram: http://instagram.com/armandohasudungan Twitter: https://twitter.com/Armando71021105 -

How to Study Biochemistry in Medical School
Link to the books in this video on Amazon: Rapid Review Biochemistry: http://www.amazon.com/gp/product/0323068871/ref=as_li_ss_tl?ie=UTF8&camp;=1789&creative;=390957&creativeASIN;=0323068871&linkCode;=as2&tag;=doctheundfor-20 Lippincott's Biochemistry http://www.amazon.com/gp/product/160831412X/ref=as_li_ss_tl?ie=UTF8&camp;=1789&creative;=390957&creativeASIN;=160831412X&linkCode;=as2&tag;=doctheundfor-20 Lehninger Principles of Biochemistry http://www.amazon.com/gp/product/071677108X/ref=as_li_ss_tl?ie=UTF8&camp;=1789&creative;=390957&creativeASIN;=071677108X&linkCode;=as2&tag;=doctheundfor-20 -

Biological Molecules - You Are What You Eat: Crash Course Biology #3
Hank talks about the molecules that make up every living thing - carbohydrates, lipids, and proteins - and how we find them in our environment and in the food that we eat. Crash Course Biology is now available on DVD! http://dftba.com/product/1av/CrashCourse-Biology-The-Complete-Series-DVD-Set Follow CrashCourse on Twitter: http://www.twitter.com/thecrashcourse Like CrashCourse on Facebook: http://www.facebook.com/youtubecrashcourse Resources for this episode in the Google Document here: http://dft.ba/-citations2 TAGS: biological molecules, carbohydrates, lipids, proteins, nucleic acids, food, biolography, william prout, urea, energy, monosaccharides, glucose, fructose, disaccharides, sucrose, polysaccharides, simple sugars, cellulose, starch, glycogen, glycerol, fatty acid, triglyc... -

Video Lecture: Introduction to Biochemistry
Introductory topics related to biochemistry. Atoms, molecules, and the basis of chemistry is discussed by Mr. Joseph Lamb -

Biochemistry 4.0: Introduction to amino acids
Includes a review of protein function, the basics of stereochemistry (L- and D-amino acids) and a run through of all amino acids found in proteins. -

Biochemistry (Molecular and Cellular) at Oxford University
Want to know more about studying at Oxford University? Watch this short film to hear tutors and students talk about this undergraduate degree. For more information on this course, please visit our website at http://www.ox.ac.uk/admissions/undergraduate_courses/courses/biochemistry_molecular_and_cellular/biochemistry.html -

#1 Biochemistry Lecture (Introduction) from Kevin Ahern's BB 350
1. Contact me at kgahern@davincipress.com / Friend me on Facebook (kevin.g.ahern) 2. Download my free biochemistry book at http://biochem.science.oregonstate.edu/biochemistry-free-and-easy 3. Take my free iTunes U course at https://itunes.apple.com/us/course/biochemistry/id556410409 4. Check out my free book for pre-meds at http://biochem.science.oregonstate.edu/biochemistry-free-and-easy 5. Lecturio videos for medical students - https://www.lecturio.com/medical-courses/biochemistry.course 6. Course video channel at http://www.youtube.com/user/oharow/videos?view=1 7. Check out all of my free workshops at http://oregonstate.edu/dept/biochem/ahern/123.html 8. Check out my Metabolic Melodies at http://www.davincipress.com/ 9. My courses can be taken for credit (wherever you live) via OSU's e... -

A day in the life of a biochemistry student
Ever wonder what the life of a science major, particularly a biochemistry student, is really like? -Biochemistry, B.S. (May 2013) California State University, Fullerton Music: Walking on Sunshine - Katrina and the Waves Carmina Burana: O Fortuna - Carl Orff Also Sprach Zarathustr - Richard Strauss Rawhide Remix - Blues Brothers Cats on Mars - Cowboy Bepop Time (Inception) - Hans Zimmer **the last scene is directly related to Inception** i.e. dreams within dreams of trying to solve puzzles representing finishing that o chem hw in real life -

CAREERS IN B.SC BIOCHEMISTRY– M.Sc,P.Hd,Research Institutes,Job Opportunities,Salary Package
CAREERS IN B.SC BIOCHEMISTRY.Go through the career opportunities of B.SC BIOCHEMISTRY, Govt jobs and Employment News channel from Freshersworld.com – The No.1 job portal for freshers in India. Visit http://www.freshersworld.com?src=Youtube for detailed Career information,Job Opportunities,Education details of B.SC BIOCHEMISTRY. Bachelors in Science in Biochemistry is an exciting venture these days. There are quite a few job openings for this field, and there are some good courses available for further studies. We shall discuss a few career opportunities available to BSc graduates and also a few options available for higher studies. Biochemistry involves the study of chemical and biological processes in organisms at a very minute level. The course offers you insights into various biologi... -

Biochemistry Basics
Biochemistry Basics -

What is biochemistry?
A brief and concise introduction to biochemistry. What is it? What does it involve? And why is it important? -

Introduction to Biochemistry Lecture by Kevin Ahern, Part 1 of 4
1. Contact me at kgahern@davincipress.com / Friend me on Facebook (kevin.g.ahern) 2. Download my free biochemistry book at http://biochem.science.oregonstate.edu/biochemistry-free-and-easy 3. Take my free iTunes U course at https://itunes.apple.com/us/course/biochemistry/id556410409 4. Check out my free book for pre-meds at http://biochem.science.oregonstate.edu/biochemistry-free-and-easy 5. Lecturio videos for medical students - https://www.lecturio.com/medical-courses/biochemistry.course 6. Course video channel at http://www.youtube.com/user/oharow/videos?view=1 7. Check out all of my free workshops at http://oregonstate.edu/dept/biochem/ahern/123.html 8. Check out my Metabolic Melodies at http://www.davincipress.com/ 9. My courses can be taken for credit (wherever you live) via OSU's e... -

BSc Biochemistry - School of Biosciences
Charlotte - an undergraduate student in the School of Biosciences - describes her experiences of the BSc Biochemistry degree course and of her time at the University of Birmingham. http://www.birmingham.ac.uk/students/courses/undergraduate/biosciences/biochemistry.aspx -

1st Lecture Kaplan Step 1 CA Biochemistry & Medical Genetics Turco Jan 6, 2014
-

Biochemistry - Memorize Glycolysis Using This Mnemonic
Glycolysis is a sequence of ten enzyme-catalyzed reactions (HPPATGPPEP), which can be remembered using the mnemonic Hungry Pirates Picked All The Greatest Pickled Pumpkins Ever Picked. -

Biochemistry; Be inspired
We hope you enjoy this student perspective of Biochemistry degree at the University of Bath and are inspired to consider this fantastic option for higher education. -

Hitler takes biochemistry
Hitler gets his biochemistry test results back -

The Difference Between Biochemistry & Chemical Biology : Chemistry Lessons
Subscribe Now: http://www.youtube.com/subscription_center?add_user=ehoweducation Watch More: http://www.youtube.com/ehoweducation The difference between biochemistry and chemical biology has to do with things like lipids and proteins. Find out about the difference between bio chemistry and chemical biology with help from an experienced chemistry professional in this free video clip. Expert: Robin Higgins Contact: www.linkedin.com/pub/robin-higgins/31/6aa/9 Bio: Robin Higgins graduated with a B.S in Chemistry from Emory University 2010, and has just recently received her M.S in Chemistry from the University of California Los Angeles. Filmmaker: bjorn wilde Series Description: Chemistry plays a very important role in all of our lives each and every day. Get tips on chemistry with help f... -

Biochemistry Water, PH and Buffers Part 1 tutorial
Biochemistry with Professor Paul M. Bingham View the full video at http://www.streamingtutors.com/ -

Biochemistry Lipids tutorial
Biochemistry with Professor Paul M. Bingham View the full video at http://www.streamingtutors.com/. -

#41 Biochemistry DNA Replication I Lecture for Kevin Ahern's BB 451/551
1. Contact me at kgahern@davincipress.com / Friend me on Facebook (kevin.g.ahern) 2. Download my free biochemistry book at http://biochem.science.oregonstate.edu/biochemistry-free-and-easy 3. Take my free iTunes U course at https://itunes.apple.com/us/course/biochemistry/id556410409 4. Check out my free book for pre-meds at http://biochem.science.oregonstate.edu/biochemistry-free-and-easy 5. Course video channel at http://www.youtube.com/user/oharow/videos?view=1 6. Check out all of my free workshops at http://www.youtube.com/playlist?list=PLlnFrNM93wqyTiCLZKehU1Tp8rNmnOWYB&feature;=view_all 7. Check out my Metabolic Melodies at http://www.davincipress.com/metabmelodies.html 8. My courses can be taken for credit (wherever you live) via OSU's ecampus. For details, see http://ecampus.oregons... -

Lecture - 1 Amino Acids I
Lecture Series on BioChemistry I by Prof.S.Dasgupta, Dept of Chemistry, IIT Kharagpur. For more details on NPTEl visit http://nptel.iitm.ac.in
Introduction to Biochemistry HD
- Order: Reorder
- Duration: 3:49
- Updated: 31 Dec 2013
- views: 167781
- published: 31 Dec 2013
- views: 167781
Biochemistry of Carbohydrates
- Order: Reorder
- Duration: 16:15
- Updated: 06 Jan 2015
- views: 85500
- published: 06 Jan 2015
- views: 85500
How to Study Biochemistry in Medical School
- Order: Reorder
- Duration: 9:12
- Updated: 05 Dec 2012
- views: 102562
- published: 05 Dec 2012
- views: 102562
Biological Molecules - You Are What You Eat: Crash Course Biology #3
- Order: Reorder
- Duration: 14:09
- Updated: 13 Feb 2012
- views: 1733264
- published: 13 Feb 2012
- views: 1733264
Video Lecture: Introduction to Biochemistry
- Order: Reorder
- Duration: 10:23
- Updated: 20 Mar 2013
- views: 33799
- published: 20 Mar 2013
- views: 33799
Biochemistry 4.0: Introduction to amino acids
- Order: Reorder
- Duration: 15:29
- Updated: 25 Jan 2015
- views: 5234
- published: 25 Jan 2015
- views: 5234
Biochemistry (Molecular and Cellular) at Oxford University
- Order: Reorder
- Duration: 12:24
- Updated: 07 Oct 2013
- views: 33909
- published: 07 Oct 2013
- views: 33909
#1 Biochemistry Lecture (Introduction) from Kevin Ahern's BB 350
- Order: Reorder
- Duration: 49:33
- Updated: 03 Apr 2013
- views: 22785
- published: 03 Apr 2013
- views: 22785
A day in the life of a biochemistry student
- Order: Reorder
- Duration: 9:18
- Updated: 14 Apr 2013
- views: 20679
- published: 14 Apr 2013
- views: 20679
CAREERS IN B.SC BIOCHEMISTRY– M.Sc,P.Hd,Research Institutes,Job Opportunities,Salary Package
- Order: Reorder
- Duration: 4:34
- Updated: 14 Jul 2015
- views: 2766
- published: 14 Jul 2015
- views: 2766
Biochemistry Basics
- Order: Reorder
- Duration: 15:02
- Updated: 21 Aug 2013
- views: 26226
What is biochemistry?
- Order: Reorder
- Duration: 5:20
- Updated: 01 Nov 2014
- views: 8023
- published: 01 Nov 2014
- views: 8023
Introduction to Biochemistry Lecture by Kevin Ahern, Part 1 of 4
- Order: Reorder
- Duration: 14:47
- Updated: 30 Aug 2010
- views: 58381
- published: 30 Aug 2010
- views: 58381
BSc Biochemistry - School of Biosciences
- Order: Reorder
- Duration: 4:03
- Updated: 26 Nov 2012
- views: 10184
- published: 26 Nov 2012
- views: 10184
1st Lecture Kaplan Step 1 CA Biochemistry & Medical Genetics Turco Jan 6, 2014
- Order: Reorder
- Duration: 256:12
- Updated: 09 Nov 2015
- views: 1672
- published: 09 Nov 2015
- views: 1672
Biochemistry - Memorize Glycolysis Using This Mnemonic
- Order: Reorder
- Duration: 9:37
- Updated: 26 Feb 2015
- views: 22535
- published: 26 Feb 2015
- views: 22535
Biochemistry; Be inspired
- Order: Reorder
- Duration: 8:29
- Updated: 19 Nov 2012
- views: 18853
- published: 19 Nov 2012
- views: 18853
Hitler takes biochemistry
- Order: Reorder
- Duration: 3:59
- Updated: 08 Nov 2011
- views: 9383
The Difference Between Biochemistry & Chemical Biology : Chemistry Lessons
- Order: Reorder
- Duration: 2:02
- Updated: 03 Mar 2014
- views: 4219
- published: 03 Mar 2014
- views: 4219
Biochemistry Water, PH and Buffers Part 1 tutorial
- Order: Reorder
- Duration: 11:16
- Updated: 15 Feb 2014
- views: 7242
- published: 15 Feb 2014
- views: 7242
Biochemistry Lipids tutorial
- Order: Reorder
- Duration: 11:16
- Updated: 15 Feb 2014
- views: 9880
- published: 15 Feb 2014
- views: 9880
#41 Biochemistry DNA Replication I Lecture for Kevin Ahern's BB 451/551
- Order: Reorder
- Duration: 49:11
- Updated: 16 Feb 2012
- views: 15786
- published: 16 Feb 2012
- views: 15786
Lecture - 1 Amino Acids I
- Order: Reorder
- Duration: 54:03
- Updated: 01 Feb 2008
- views: 490143
- published: 01 Feb 2008
- views: 490143
- Playlist
- Chat
- Playlist
- Chat

Introduction to Biochemistry HD
- Report rights infringement
- published: 31 Dec 2013
- views: 167781

Biochemistry of Carbohydrates
- Report rights infringement
- published: 06 Jan 2015
- views: 85500

How to Study Biochemistry in Medical School
- Report rights infringement
- published: 05 Dec 2012
- views: 102562

Biological Molecules - You Are What You Eat: Crash Course Biology #3
- Report rights infringement
- published: 13 Feb 2012
- views: 1733264

Video Lecture: Introduction to Biochemistry
- Report rights infringement
- published: 20 Mar 2013
- views: 33799

Biochemistry 4.0: Introduction to amino acids
- Report rights infringement
- published: 25 Jan 2015
- views: 5234

Biochemistry (Molecular and Cellular) at Oxford University
- Report rights infringement
- published: 07 Oct 2013
- views: 33909

#1 Biochemistry Lecture (Introduction) from Kevin Ahern's BB 350
- Report rights infringement
- published: 03 Apr 2013
- views: 22785

A day in the life of a biochemistry student
- Report rights infringement
- published: 14 Apr 2013
- views: 20679

CAREERS IN B.SC BIOCHEMISTRY– M.Sc,P.Hd,Research Institutes,Job Opportunities,Salary Package
- Report rights infringement
- published: 14 Jul 2015
- views: 2766

Biochemistry Basics
- Report rights infringement
- published: 21 Aug 2013
- views: 26226

What is biochemistry?
- Report rights infringement
- published: 01 Nov 2014
- views: 8023

Introduction to Biochemistry Lecture by Kevin Ahern, Part 1 of 4
- Report rights infringement
- published: 30 Aug 2010
- views: 58381

BSc Biochemistry - School of Biosciences
- Report rights infringement
- published: 26 Nov 2012
- views: 10184
-
Lyrics list:text lyricsplay full screenplay karaoke
Despite criticism, European Union plans are ready to deport refugees
Edit The Malta Independent 02 Apr 2016Google's April Fools' joke went so wrong that it might have cost someone their job
Edit TechRadar 01 Apr 2016IS ‘madmen’ will certainly use nukes: Obama
Edit Dawn 02 Apr 2016WN.com Week In Review For March 27-April 2, 2016
Edit WorldNews.com 01 Apr 2016Inequality Even Before Birth And After Death: How The Koch Empire Impacted Me
Edit WorldNews.com 01 Apr 2016Student Austin Weigle earns national recognition (Southern Illinois University System)
Edit Public Technologies 02 Apr 2016President Erdoğan Meets with US Opinion Leaders (Presidency of the Republic of Turkey)
Edit Public Technologies 02 Apr 2016Emily Engebretson to present master’s flute recital on Saturday, April 9 (Evangel University)
Edit Public Technologies 02 Apr 2016SIUE’s De Meo Receives Paul Simon Outstanding Teacher-Scholar Award (Southern Illinois University System)
Edit Public Technologies 02 Apr 2016For the Record, April 1, 2016 (University of Delaware)
Edit Public Technologies 01 Apr 2016Research reveals the anticancer activity of rosemary (Università di Pisa)
Edit Public Technologies 01 Apr 2016Venomous varmint could offer ways to deal with cancer
Edit The Irish Times 01 Apr 2016Lehman Alumnus at Texas A&M; Wins Award as Outstanding Academic Advisor (CUNY - The City University of New York)
Edit Public Technologies 01 Apr 2016The Right Chemistry: Au+H2O Times 2 (North Carolina State University)
Edit Public Technologies 01 Apr 2016Undergraduate research on display at MSU (Michigan State University)
Edit Public Technologies 01 Apr 2016English student tops podium in research presentation competition (University of Windsor)
Edit Public Technologies 01 Apr 2016UNM undergrads among first to train using new genome editing technology (The University of New Mexico)
Edit Public Technologies 01 Apr 2016SIUE’s De Meo Receives Paul Simon Outstanding Teacher-Scholar Award (Southern Illinois University System - Edwardsville)
Edit Public Technologies 01 Apr 2016- 1
- 2
- 3
- 4
- 5
- Next page »







