- published: 20 Apr 2014
- views: 470310
-
remove the playlistPcr
-
remove the playlistLatest Videos
-
remove the playlistLongest Videos
- remove the playlistPcr
- remove the playlistLatest Videos
- remove the playlistLongest Videos
- published: 25 Mar 2015
- views: 44258
- published: 20 Jan 2015
- views: 21303
- published: 22 Mar 2010
- views: 483263
- published: 16 Apr 2009
- views: 444274
- published: 08 Dec 2014
- views: 167402
- published: 09 May 2012
- views: 141328
- published: 02 Jun 2012
- views: 214715
- published: 28 Feb 2013
- views: 74134
- published: 27 Oct 2013
- views: 62960
- published: 08 Jan 2008
- views: 1641093

The polymerase chain reaction (PCR) is a scientific technique in molecular biology to amplify a single or a few copies of a piece of DNA across several orders of magnitude, generating thousands to millions of copies of a particular DNA sequence.
Developed in 1983 by Kary Mullis, PCR is now a common and often indispensable technique used in medical and biological research labs for a variety of applications. These include DNA cloning for sequencing, DNA-based phylogeny, or functional analysis of genes; the diagnosis of hereditary diseases; the identification of genetic fingerprints (used in forensic sciences and paternity testing); and the detection and diagnosis of infectious diseases. In 1993, Mullis was awarded the Nobel Prize in Chemistry along with Michael Smith for his work on PCR.
The method relies on thermal cycling, consisting of cycles of repeated heating and cooling of the reaction for DNA melting and enzymatic replication of the DNA. Primers (short DNA fragments) containing sequences complementary to the target region along with a DNA polymerase (after which the method is named) are key components to enable selective and repeated amplification. As PCR progresses, the DNA generated is itself used as a template for replication, setting in motion a chain reaction in which the DNA template is exponentially amplified. PCR can be extensively modified to perform a wide array of genetic manipulations.
This article is licensed under the Creative Commons Attribution-ShareAlike 3.0 Unported License, which means that you can copy and modify it as long as the entire work (including additions) remains under this license.
- Loading...

-
 3:56
3:56PCR (Polymerase Chain Reaction)
PCR (Polymerase Chain Reaction)PCR (Polymerase Chain Reaction)
PCR technique (Polymerase Chain Reaction), Animation. It is a technique used to make multiple copies of a segment DNA of interest, generating a large amount of copies from a small initial simple. Amplification of DNA segments makes possible the detection of pathogenic virus or bacteria, identification of individuals (DNA fingerprinting), and several scientific research involving DNA manipulation. Spanish version: http://youtu.be/TalHTjA5gKU This video has been produced in the Institute of General Organic Chemistry of the CSIC (IQOG-CSIC) by Guillermo Corrales, as part of its task for promoting Science Communication and may be freely used for educational and science popularization purposes Instituto de Química Orgánica General (QOG) CSIC http://www.youtube.com/user/CanalDivulgacion -
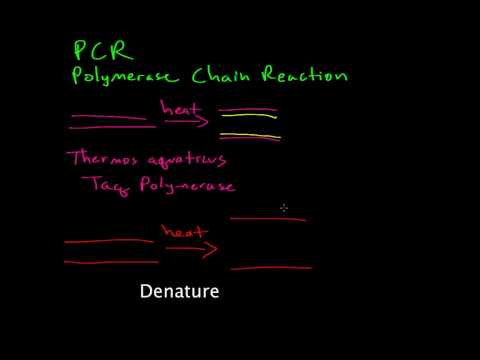 4:38
4:38Polymerase chain reaction (PCR)
Polymerase chain reaction (PCR)Polymerase chain reaction (PCR)
Visit us (http://www.khanacademy.org/science/healthcare-and-medicine) for health and medicine content or (http://www.khanacademy.org/test-prep/mcat) for MCAT related content. These videos do not provide medical advice and are for informational purposes only. The videos are not intended to be a substitute for professional medical advice, diagnosis or treatment. Always seek the advice of a qualified health provider with any questions you may have regarding a medical condition. Never disregard professional medical advice or delay in seeking it because of something you have read or seen in any Khan Academy video. -
 13:14
13:14Polymerase Chain Reaction (PCR)
Polymerase Chain Reaction (PCR)Polymerase Chain Reaction (PCR)
Donate here: http://www.aklectures.com/donate.php Website video link: http://www.aklectures.com/lecture/polymerase-chain-reaction-pcr Facebook link: https://www.facebook.com/aklectures Website link: http://www.aklectures.com -
 1:28
1:28Polymerase Chain Reaction (PCR)
Polymerase Chain Reaction (PCR)Polymerase Chain Reaction (PCR)
Polymerase chain reaction (PCR) allows researchers to amplify DNA in a test tube. This process uses an enzyme derived from heat-resistant bacteria. The steps of PCR are driven by changes in temperature. Originally created for DNA Interactive ( http://www.dnai.org ). TRANSCRIPT: Polymerase chain reaction (PCR) is a process where many copies of a specific piece of DNA can be made. This is known as amplification. Double-stranded DNA (red) unwinds and separates when the temperature is increased. As the temperature is decreased, small starter sequences called primers (glowing) can attach or anneal to the DNA. These primer sequences are usually only 20 to 25 nucleotides long, and are designed to match the start and end points of the DNA piece to be amplified. Once the primers have annealed, Taq polymerase (blue) copies the DNA starting from the primer. The temperature is increased; the strands separate; more primers anneal; the DNA is copied; and this cycle is repeated many times. In a typical PCR reaction there are 30 cycles, which can potentially create one billion copies starting from one molecule of DNA. -
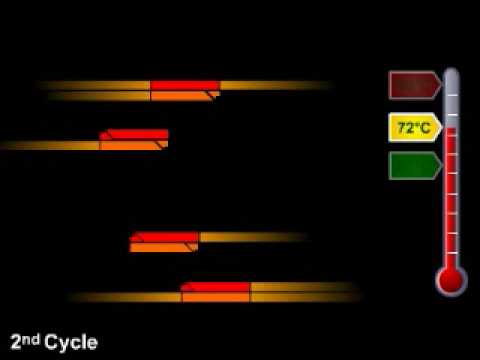 2:53
2:53Polymerase Chain Reaction (PCR)
Polymerase Chain Reaction (PCR)Polymerase Chain Reaction (PCR)
To purchase the book: http://www.garlandscience.com/product/isbn/9780815344544 To see more videos like this, please visit: http://www.garlandscience.com/garlandscience_resources/book_resources.jsf?chapter=ALL_CHAPTERS&selectedPage;=1&landing;=student&resultsPerPage;=10&isbn;=9780815344544&tabId;=ALL_RESOURCES&conversationId;=321516 The polymerase chain reaction, or PCR, amplifies a specific DNA fragment from a complex mixture. This video explains the process. This video is from: Essential Cell Biology, 4th Edition Alberts, Bray, Hopkin, Johnson, Lewis, Raff, Roberts, & Walter ISBN: 978-0-8153-4454-4 -
 6:05
6:05Die PCR-Methode einfach erklärt
Die PCR-Methode einfach erklärtDie PCR-Methode einfach erklärt
Hier seht ihr, was die PCR-Methode ist und wie sie funktioniert. Wir erklären euch jeden einzelnen Schritt und zeigen euch, warum diese Methode so praktisch ist! Kategorie: Abitur Hier kommst du direkt zum nächsten Video: https://www.youtube.com/watch?v=XmKNN5oePGo&index;=1&list;=PLtB1MJSQL5hRsPZTFo3tRPvHgtoFEM7fb » ALLE KANÄLE Wirtschaft: http://www.thesimpleeconomics.de Mathe: https://www.youtube.com/TheSimpleMaths Biologie: http://www.thesimplebiology.de Physik: http://www.thesimplephysics.de Chemie: http://www.thesimplechemics.de » MEHR VON UNS Twitter: http://www.twitter.com/thesimpleclub Facebook: http://fb.thesimpleclub.de Alex auf Instagram: http://alex.thesimpleclub.de Nico auf Instagram: http://nico.thesimpleclub.de » WAS IST THE SIMPLE CLUB? Wir sind der Meinung, dass Bildung Spaß machen muss. Deswegen bieten wir dir auf 4 Kanälen die beste und unterhaltsamste Nachhilfe die du im Netz finden kannst: Und das in Mathematik, Biologie, Chemie und Physik. In verschiedenen Kategorien und Schwierigkeitsgraden bereiten wir dich auf deine Prüfung vor. Egal ob Schüler oder Student, ob jung oder alt, bei uns findet jeder die passenden Videos. Und das Beste: TheSimpleClub ist und bleibt komplett kostenlos! » CREDITS Ein Konzept von Alexander Giesecke und Nicolai Schork Geschrieben von: Félix Selinger Visuelle Konzeption: Antje Heidemann Ton: Alexander Giesecke Schnitt & Effekte: Vincent Viebig » BILDQUELLEN Prezi-Bilder: Thermocycler Baby Blue: Science Museum London / Science and Society Picture Library - 'Baby Blue' - a prototype polymerase chain reaction (PCR), c 1986 (Wikimedia Commons), http://bit.ly/1v2TYB5, CC BY-SA 2.0 Eppendorf-Gefäß: Nadina Wiórkiewicz (Nadine90) (Wikimedia Commons), http://bit.ly/1v33PGi, CC BY-SA 3.0 -
 8:35
8:35Polymerase Chain Reaction (PCR) | MIT 7.01SC Fundamentals of Biology
Polymerase Chain Reaction (PCR) | MIT 7.01SC Fundamentals of BiologyPolymerase Chain Reaction (PCR) | MIT 7.01SC Fundamentals of Biology
Polymerase Chain Reaction (PCR) Instructor: Robert Dorkin View the complete course: http://ocw.mit.edu/7-01SCF11 License: Creative Commons BY-NC-SA More information at http://ocw.mit.edu/terms More courses at http://ocw.mit.edu -
 3:32
3:32PCR - Polymerase Chain Reaction - Simple Animated Tutorial
PCR - Polymerase Chain Reaction - Simple Animated TutorialPCR - Polymerase Chain Reaction - Simple Animated Tutorial
This is a very basic video about PCR. Pause if it goes too fast. Please subscribe=) PCR problems- What are primers dimers? https://www.youtube.com/watch?v=hyqORD3YmNE In more detail: The role of Magnesium Chloride in PCR https://www.youtube.com/watch?v=4b1B_Ct43rw QUick facts on Taq polymerase https://www.youtube.com/watch?v=NdalZQteXj0 Real Time PCR http://youtu.be/EaGH1eKfvC0 -
 4:38
4:38PCR
PCRPCR
PCR -
 3:42
3:42How to Set Up the PCR Reaction
How to Set Up the PCR ReactionHow to Set Up the PCR Reaction
Learn more at http://www.lifetechnologies.com/pcr This video is the second part of a 3-part Getting started with PCR series that shows how to set up and run a PCR experiment. The success and failure of any PCR experiment is dependent on preparation. This particular video shows the different components that go into a PCR reaction, how each component contributes to the experiment, and lastly, when to set up a master mix. -
 30:25
30:25PCR (polymerase chain reaction) in detail
PCR (polymerase chain reaction) in detailPCR (polymerase chain reaction) in detail
This pcr lecture explains polymerase chain reaction in details. Web- http://shomusbiology.weebly.com/ Download the study materials here- http://shomusbiology.weebly.com/bio-materials.html The polymerase chain reaction (PCR) is a biochemical technology in molecular biology to amplify a single or a few copies of a piece of DNA across several orders of magnitude, generating thousands to millions of copies of a particular DNA sequence. Developed in 1983 by Kary Mullis,[1][2] PCR is now a common and often indispensable technique used in medical and biological research labs for a variety of applications.[3][4] These include DNA cloning for sequencing, DNA-based phylogeny, or functional analysis of genes; the diagnosis of hereditary diseases; the identification of genetic fingerprints (used in forensic sciences and paternity testing); and the detection and diagnosis of infectious diseases. In 1993, Mullis was awarded the Nobel Prize in Chemistry along with Michael Smith for his work on PCR.[5] The method relies on thermal cycling, consisting of cycles of repeated heating and cooling of the reaction for DNA melting and enzymatic replication of the DNA. Primers (short DNA fragments) containing sequences complementary to the target region along with a DNA polymerase (after which the method is named) are key components to enable selective and repeated amplification. As PCR progresses, the DNA generated is itself used as a template for replication, setting in motion a chain reaction in which the DNA template is exponentially amplified. PCR can be extensively modified to perform a wide array of genetic manipulations. Almost all PCR applications employ a heat-stable DNA polymerase, such as Taq polymerase, an enzyme originally isolated from the bacterium Thermus aquaticus. This DNA polymerase enzymatically assembles a new DNA strand from DNA building-blocks, the nucleotides, by using single-stranded DNA as a template and DNA oligonucleotides (also called DNA primers), which are required for initiation of DNA synthesis. The vast majority of PCR methods use thermal cycling, i.e., alternately heating and cooling the PCR sample through a defined series of temperature steps. In the first step, the two strands of the DNA double helix are physically separated at a high temperature in a process called DNA melting. In the second step, the temperature is lowered and the two DNA strands become templates for DNA polymerase to selectively amplify the target DNA. The selectivity of PCR results from the use of primers that are complementary to the DNA region targeted for amplification under specific thermal cycling conditions. Source of the article published in description is Wikipedia. I am sharing their material. Copyright by original content developers of Wikipedia. Link- http://en.wikipedia.org/wiki/Main_Page -
 2:14
2:14The PCR Song
The PCR SongThe PCR Song
"Scientists for Better PCR" a Bio-Rad Music Video for the all new 1000-Series Thermal Cyclers Click below for more information: http://bio-rad.cnpg.com/lsca/videos/ScientistsForBetterPCR/ -
 8:56
8:56Aula de PCR e eletroforese
Aula de PCR e eletroforeseAula de PCR e eletroforese
-
 11:55
11:55PCR for A-level
PCR for A-levelPCR for A-level
The Polymerase Chain Reaction (PCR) and its use in scientific research PCR, which amplifies short sections of DNA, is used in many areas of the biological sciences. Final year B. Sc. Biological Sciences student William Fraser chose to produce a video about PCR and its use to detect bacteria without the need to grow them in the laboratory. Working with staff at Sir John Deane's Sixth Form College, Northwich, Cheshire and research staff from the Institute of integrative Biology at the University of Liverpool, he planned, filmed and edited the short video. It starts with an animation of the steps in PCR (targeted at A-level requirements) and then shows Dr David Rooks using PCR in research about the microorganisms that breakdown cellulose.
- Allele
- Allele-specific PCR
- Amplicon
- Anaerobic organism
- Animal model
- Annealing (biology)
- Antibody
- Applications of PCR
- Arithmetic
- Asymmetric PCR
- Biotechnology
- Buffer solution
- Cations
- CDNA
- Cetus Corporation
- Chain reaction
- Complementary DNA
- Divalent
- DNA
- DNA cloning
- DNA double helix
- DNA footprinting
- DNA helicase
- DNA ladder
- DNA melting
- DNA polymerase
- DNA replication
- DNA sequence
- DNA sequencing
- DuPont
- Emeryville
- Enzyme
- Escherichia coli
- Exons
- Exponential growth
- Expression profiling
- Fluorophore
- Forensic analysis
- Forensic science
- Formamide
- Gel electrophoresis
- Gene
- Gene expression
- Genetic engineering
- Genetic fingerprint
- Genome
- Genome walking
- Genomic
- Hereditary disease
- Hoffmann-La Roche
- Hot start PCR
- Hybridization probe
- Hydroxyl group
- In silico PCR
- In vitro
- Infectious disease
- Introns
- Kary Mullis
- Kilo base pair
- Leukemia
- Lymphoma
- Magnesium
- Malignant
- Mammoth
- Manganese
- Miniprimer PCR
- Molecular biology
- Multiplex-PCR
- Mummy
- Mycobacterium
- Nested PCR
- Northern blot
- Nucleotide
- Oligonucleotide
- PAN-AC
- Parental testing
- Paul Rabinow
- PCR (disambiguation)
- PCR mutagenesis
- PCR optimization
- Phosphate group
- Phylogeny
- Plasmid
- Polony
- Potassium
- Promega
- PubMed Central
- PubMed Identifier
- Q-PCR
- RACE (biology)
- Real-time PCR
- Recombinant DNA
- Restriction digest
- RNA
- RT-PCR
- Russia
- Scientific American
- Scientific technique
- Self ligation
- Sequencing
- SNP genotyping
- Solid Phase PCR
- Southern blot
- TAIL-PCR
- Taq polymerase
- TaqMan
- Template PCR
- Template talk PCR
- Thermal cycler
- Thermocycler
- Thermophile
- Thermus aquaticus
- Tissue culture
- Touchdown PCR
- Transcriptome
- Tsar
- Variants of PCR
- Viral load
- Virus
-

PCR (Polymerase Chain Reaction)
PCR technique (Polymerase Chain Reaction), Animation. It is a technique used to make multiple copies of a segment DNA of interest, generating a large amount of copies from a small initial simple. Amplification of DNA segments makes possible the detection of pathogenic virus or bacteria, identification of individuals (DNA fingerprinting), and several scientific research involving DNA manipulation. Spanish version: http://youtu.be/TalHTjA5gKU This video has been produced in the Institute of General Organic Chemistry of the CSIC (IQOG-CSIC) by Guillermo Corrales, as part of its task for promoting Science Communication and may be freely used for educational and science popularization purposes Instituto de Química Orgánica General (QOG) CSIC http://www.youtube.com/user/CanalDivulgacion -

Polymerase chain reaction (PCR)
Visit us (http://www.khanacademy.org/science/healthcare-and-medicine) for health and medicine content or (http://www.khanacademy.org/test-prep/mcat) for MCAT related content. These videos do not provide medical advice and are for informational purposes only. The videos are not intended to be a substitute for professional medical advice, diagnosis or treatment. Always seek the advice of a qualified health provider with any questions you may have regarding a medical condition. Never disregard professional medical advice or delay in seeking it because of something you have read or seen in any Khan Academy video. -

Polymerase Chain Reaction (PCR)
Donate here: http://www.aklectures.com/donate.php Website video link: http://www.aklectures.com/lecture/polymerase-chain-reaction-pcr Facebook link: https://www.facebook.com/aklectures Website link: http://www.aklectures.com -

Polymerase Chain Reaction (PCR)
Polymerase chain reaction (PCR) allows researchers to amplify DNA in a test tube. This process uses an enzyme derived from heat-resistant bacteria. The steps of PCR are driven by changes in temperature. Originally created for DNA Interactive ( http://www.dnai.org ). TRANSCRIPT: Polymerase chain reaction (PCR) is a process where many copies of a specific piece of DNA can be made. This is known as amplification. Double-stranded DNA (red) unwinds and separates when the temperature is increased. As the temperature is decreased, small starter sequences called primers (glowing) can attach or anneal to the DNA. These primer sequences are usually only 20 to 25 nucleotides long, and are designed to match the start and end points of the DNA piece to be amplified. Once the primers have annealed, T... -

Polymerase Chain Reaction (PCR)
To purchase the book: http://www.garlandscience.com/product/isbn/9780815344544 To see more videos like this, please visit: http://www.garlandscience.com/garlandscience_resources/book_resources.jsf?chapter=ALL_CHAPTERS&selectedPage;=1&landing;=student&resultsPerPage;=10&isbn;=9780815344544&tabId;=ALL_RESOURCES&conversationId;=321516 The polymerase chain reaction, or PCR, amplifies a specific DNA fragment from a complex mixture. This video explains the process. This video is from: Essential Cell Biology, 4th Edition Alberts, Bray, Hopkin, Johnson, Lewis, Raff, Roberts, & Walter ISBN: 978-0-8153-4454-4 -

Die PCR-Methode einfach erklärt
Hier seht ihr, was die PCR-Methode ist und wie sie funktioniert. Wir erklären euch jeden einzelnen Schritt und zeigen euch, warum diese Methode so praktisch ist! Kategorie: Abitur Hier kommst du direkt zum nächsten Video: https://www.youtube.com/watch?v=XmKNN5oePGo&index;=1&list;=PLtB1MJSQL5hRsPZTFo3tRPvHgtoFEM7fb » ALLE KANÄLE Wirtschaft: http://www.thesimpleeconomics.de Mathe: https://www.youtube.com/TheSimpleMaths Biologie: http://www.thesimplebiology.de Physik: http://www.thesimplephysics.de Chemie: http://www.thesimplechemics.de » MEHR VON UNS Twitter: http://www.twitter.com/thesimpleclub Facebook: http://fb.thesimpleclub.de Alex auf Instagram: http://alex.thesimpleclub.de Nico auf Instagram: http://nico.thesimpleclub.de » WAS IST THE SIMPLE CLUB? Wir sind der Meinung, dass Bil... -

Polymerase Chain Reaction (PCR) | MIT 7.01SC Fundamentals of Biology
Polymerase Chain Reaction (PCR) Instructor: Robert Dorkin View the complete course: http://ocw.mit.edu/7-01SCF11 License: Creative Commons BY-NC-SA More information at http://ocw.mit.edu/terms More courses at http://ocw.mit.edu -

PCR - Polymerase Chain Reaction - Simple Animated Tutorial
This is a very basic video about PCR. Pause if it goes too fast. Please subscribe=) PCR problems- What are primers dimers? https://www.youtube.com/watch?v=hyqORD3YmNE In more detail: The role of Magnesium Chloride in PCR https://www.youtube.com/watch?v=4b1B_Ct43rw QUick facts on Taq polymerase https://www.youtube.com/watch?v=NdalZQteXj0 Real Time PCR http://youtu.be/EaGH1eKfvC0 -

PCR
PCR -

How to Set Up the PCR Reaction
Learn more at http://www.lifetechnologies.com/pcr This video is the second part of a 3-part Getting started with PCR series that shows how to set up and run a PCR experiment. The success and failure of any PCR experiment is dependent on preparation. This particular video shows the different components that go into a PCR reaction, how each component contributes to the experiment, and lastly, when to set up a master mix. -

PCR (polymerase chain reaction) in detail
This pcr lecture explains polymerase chain reaction in details. Web- http://shomusbiology.weebly.com/ Download the study materials here- http://shomusbiology.weebly.com/bio-materials.html The polymerase chain reaction (PCR) is a biochemical technology in molecular biology to amplify a single or a few copies of a piece of DNA across several orders of magnitude, generating thousands to millions of copies of a particular DNA sequence. Developed in 1983 by Kary Mullis,[1][2] PCR is now a common and often indispensable technique used in medical and biological research labs for a variety of applications.[3][4] These include DNA cloning for sequencing, DNA-based phylogeny, or functional analysis of genes; the diagnosis of hereditary diseases; the identification of genetic fingerprints (used in ... -

The PCR Song
"Scientists for Better PCR" a Bio-Rad Music Video for the all new 1000-Series Thermal Cyclers Click below for more information: http://bio-rad.cnpg.com/lsca/videos/ScientistsForBetterPCR/ -

Aula de PCR e eletroforese
-

PCR for A-level
The Polymerase Chain Reaction (PCR) and its use in scientific research PCR, which amplifies short sections of DNA, is used in many areas of the biological sciences. Final year B. Sc. Biological Sciences student William Fraser chose to produce a video about PCR and its use to detect bacteria without the need to grow them in the laboratory. Working with staff at Sir John Deane's Sixth Form College, Northwich, Cheshire and research staff from the Institute of integrative Biology at the University of Liverpool, he planned, filmed and edited the short video. It starts with an animation of the steps in PCR (targeted at A-level requirements) and then shows Dr David Rooks using PCR in research about the microorganisms that breakdown cellulose. -

Agarose Gel Electrophoresis of DNA fragments amplified using PCR
This video is the third lesson in a series of resources detailing the PCR process and surrounding activities. It shows how to analyse a DNA sample using agarose gel electrophoresis, as well as how to make the agarose gel. -

PCR types
This PCR lecture explains about different types of PCR like nested PCR, realtime PCR, quantitative PCR, multiplex PCR, hot start PCR. It explains the principle of polymerase chain reaction techniques in details. http://shomusbiology.weebly.com/ Download the study materials here- http://shomusbiology.weebly.com/bio-materials.html Assembly PCR or Polymerase Cycling Assembly (PCA): artificial synthesis of long DNA sequences by performing PCR on a pool of long oligonucleotides with short overlapping segments. The oligonucleotides alternate between sense and antisense directions, and the overlapping segments determine the order of the PCR fragments, thereby selectively producing the final long DNA product.[23] Asymmetric PCR: preferentially amplifies one DNA strand in a double-stranded DNA tem... -

Biologie online lernen: Polymerase-Kettenreaktion (PCR)
Abiturvorbereitung in Biologie unter https://www.abiweb.de/abitur-online-lernen/biologie Die PCR (Polymerase-Ketten-Reaktion) ist eine sehr gute Methode DNA im Labor zu vervielfältigen. Aus einer DNA-Vorlage (Template) können innerhalb von 30 PCR-Cyclen mehr als 1 Milliarde Kopien dieses bestimmten DNA-Stücks erzeugt werden. Mehr Informationen zum Thema unter https://www.abiweb.de/biologie-molekularbiologie-genetik/methoden-der-gen-und-reproduktionstechnik/methode-polymerase-ketten-reaktion.html -

-

-

أساسيات تفاعل الـ PCR
شرح مفصل لخطوات تفاعل الـ PCR مع عرض مثال لعزل جين محدد. -

Real time PCR
This pcr reaction lecture explains about real time pcr procedure. It explains the realtime pcr mechanism and uses in molecular diagnosis. Web-http://shomusbiology.com/ Download the study materials here- http://shomusbiology.weebly.com/bio-materials.html A quantitative polymerase chain reaction (qPCR), also called real-time polymerase chain reaction, is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR), which is used to amplify and simultaneously quantify a targeted DNA molecule. For one or more specific sequences in a DNA sample, quantitative PCR enables both detection and quantification. The quantity can be either an absolute number of copies or a relative amount when normalized to DNA input or additional normalizing genes. The procedure follows the... -

P.C.R.Evo [5] - Evolução é "só" uma teoria? Por que não é lei?
Se evolução fosse verdade seria lei, não teoria. Será que essa frase faz sentido? Já dianto que não. Meu Twitter: @Pirulla25 Minha fanpage: https://www.facebook.com/CanalDoPirula Vídeo do Yuri (Eu,Ciência): https://www.youtube.com/watch?v=ga9K9Kt8u1M Vídeo do Nerdologia: https://www.youtube.com/watch?v=kyGu9lTr_jM Vídeo do Papo de Primata: https://www.youtube.com/watch?v=lKpAy7xylAs Meu vídeo sobre Falseabilidade: https://www.youtube.com/watch?v=GJcvAFDW4k4 Porque eu não acredito em Evolução: https://www.youtube.com/watch?v=7vWGAun9Lm4 Demais vídeos da série: https://www.youtube.com/watch?v=LBjDKpml85c&list;=PLdlKx3uNkxdGTEVXoey0rpXkm_JHd29uC&ab;_channel=CanaldoPirula Vídeo do Slow: https://www.youtube.com/watch?v=mcVKhZN9BOM Macaco Aquático: https://en.wikipedia.org/wiki/Aquatic_ap... -

Conheça os 4 ritmos de PCR e como identificá-los | SBV | MDB | IBRAPH
Saiba como se tornar um Instrutor de SBV Credenciado, Baixe Seu E-book Gratuito agora. Acesse: http://ibraph.com.br/sbv-mdb-2016/ A palestra que lhe ensina de uma forma diferente os 4 ritmos de PCR e como identificá-los. Além disso, abordo também o que fazer de forma resumida no SBV. http://youtu.be/NevV6W2fNZw
PCR (Polymerase Chain Reaction)
- Order: Reorder
- Duration: 3:56
- Updated: 20 Apr 2014
- views: 470310
- published: 20 Apr 2014
- views: 470310
Polymerase chain reaction (PCR)
- Order: Reorder
- Duration: 4:38
- Updated: 25 Mar 2015
- views: 44258
- published: 25 Mar 2015
- views: 44258
Polymerase Chain Reaction (PCR)
- Order: Reorder
- Duration: 13:14
- Updated: 20 Jan 2015
- views: 21303
- published: 20 Jan 2015
- views: 21303
Polymerase Chain Reaction (PCR)
- Order: Reorder
- Duration: 1:28
- Updated: 22 Mar 2010
- views: 483263
- published: 22 Mar 2010
- views: 483263
Polymerase Chain Reaction (PCR)
- Order: Reorder
- Duration: 2:53
- Updated: 16 Apr 2009
- views: 444274
- published: 16 Apr 2009
- views: 444274
Die PCR-Methode einfach erklärt
- Order: Reorder
- Duration: 6:05
- Updated: 08 Dec 2014
- views: 167402
- published: 08 Dec 2014
- views: 167402
Polymerase Chain Reaction (PCR) | MIT 7.01SC Fundamentals of Biology
- Order: Reorder
- Duration: 8:35
- Updated: 09 May 2012
- views: 141328
- published: 09 May 2012
- views: 141328
PCR - Polymerase Chain Reaction - Simple Animated Tutorial
- Order: Reorder
- Duration: 3:32
- Updated: 02 Jun 2012
- views: 214715
- published: 02 Jun 2012
- views: 214715
How to Set Up the PCR Reaction
- Order: Reorder
- Duration: 3:42
- Updated: 28 Feb 2013
- views: 74134
- published: 28 Feb 2013
- views: 74134
PCR (polymerase chain reaction) in detail
- Order: Reorder
- Duration: 30:25
- Updated: 27 Oct 2013
- views: 62960
- published: 27 Oct 2013
- views: 62960
The PCR Song
- Order: Reorder
- Duration: 2:14
- Updated: 08 Jan 2008
- views: 1641093
- published: 08 Jan 2008
- views: 1641093
Aula de PCR e eletroforese
- Order: Reorder
- Duration: 8:56
- Updated: 16 Oct 2014
- views: 27246
- published: 16 Oct 2014
- views: 27246
PCR for A-level
- Order: Reorder
- Duration: 11:55
- Updated: 16 May 2012
- views: 45032
- published: 16 May 2012
- views: 45032
Agarose Gel Electrophoresis of DNA fragments amplified using PCR
- Order: Reorder
- Duration: 7:43
- Updated: 29 Mar 2015
- views: 5816
- published: 29 Mar 2015
- views: 5816
PCR types
- Order: Reorder
- Duration: 42:54
- Updated: 27 Oct 2013
- views: 35150
- published: 27 Oct 2013
- views: 35150
Biologie online lernen: Polymerase-Kettenreaktion (PCR)
- Order: Reorder
- Duration: 9:32
- Updated: 03 Feb 2012
- views: 104542
- published: 03 Feb 2012
- views: 104542
Curso de Biologia Molecular - Aula 2 - PCR
- Order: Reorder
- Duration: 4:09
- Updated: 07 Oct 2013
- views: 23542
Primer Design for PCR
- Order: Reorder
- Duration: 16:15
- Updated: 03 May 2013
- views: 128115
أساسيات تفاعل الـ PCR
- Order: Reorder
- Duration: 16:00
- Updated: 07 Dec 2012
- views: 11171
Real time PCR
- Order: Reorder
- Duration: 43:43
- Updated: 25 Oct 2013
- views: 82135
- published: 25 Oct 2013
- views: 82135
P.C.R.Evo [5] - Evolução é "só" uma teoria? Por que não é lei?
- Order: Reorder
- Duration: 24:39
- Updated: 15 Mar 2016
- views: 13931
- published: 15 Mar 2016
- views: 13931
Conheça os 4 ritmos de PCR e como identificá-los | SBV | MDB | IBRAPH
- Order: Reorder
- Duration: 32:43
- Updated: 01 Jul 2014
- views: 32235
- published: 01 Jul 2014
- views: 32235
-

-

Suporte basico de vida PCR ( Parada Cardio Respiratória) dos alunos do V termo de Enfermagem -Ajes
Trabalho -

PCR: College
The second video about Post-Secondary Career Readiness. -

Reportage PCR Vétérinaire Scanelis
Scanelis est une entreprise spécialisée dans le diagnostic vétérinaire moléculaire fut le premier laboratoire à offrir des tests de PCR quantitative (PCR temps réel) en routine pour la détection, la quantification et le typage d’agents pathogènes chez l’animal. Philippe Baralon, Président de cette structure, présente son activité et son innovation dans le Mag de l'Economie. -

PCR van driver dies in accident
Odishatv is the leading new channel in Odisha. -

TEST DU CANON-PCR #1 |Clash royale
Yoooo je vous retrouvé pour le premier épisode des PCR=Présentation Clash Royale Dans ces épisode je resterai pour vous des troupes ! Dites moi quel troupe vous voulez voir dans le prochain épisode PEACE -

BIGUP-PCR-2404
BIGUPXPCRXCMWC2404 -

-

-

-

Casio PCR-T273 Electronic Cash Register
This is my personal opinion on the product casio pcr-t273 electronic cash register. You can get more information on Amazon-US: http://bit.ly/1UCHH1S Purchased this a few months ago to use during the school year to help my student learn how to make change using a "real" The product is shoddy. The don't turn, which means that program the or open the drawer. The instructions are nonsense. What a waste. Would return it but the time expired. Disappointing. CC Attribution: https://www.youtube.com/watch?v=96qjcfK44dc -

forensic files s7e12 - PCR
-

Portable and Efficient PCR Machine
Time-lapse video of PCR Machine building procedure used for demonstrations by the Langara College Bioinformatics Club. Original credit to Roshan Noronha. Editing and filming support by Jonathan Lee for Langara Bioinformatics. Music: maxzwell - Lana del Rey Blue Jeans remix Check out his music: https://soundcloud.com/maxzwell © 2016 Notorious Productions. -

PCR - Burgh Boy (Prod. By Big Jus) [explicit]
Shot by @billmikepgh & @Eastsidepopfilm -

Project CARS. PCR:A 2016 Экзаменационная гонка - LIVE (03-04-2016)
Прямая трансляция экзаменационной гонки академии WMDCARS.ru - выпуск "январь-март 2016". Трасса Oulton Park International, дистанция - 33 круга, полный суточный цикл с переменными погодными условиями. Если хотите учиться вместе с нами, то добро пожаловать на наш портал - http://wmdcars.ru -

Polymerase Chain Reaction(PCR)-Understanding the steps through PCR machine
PCR is a basic, common but a very useful technique used in research in biological science. It allows researchers to amplify a DNA sequence (or gene) of interest..... -

Reacción de PCR
Quinta sesión del proyecto de investigación PIIISA 2016 "Biofertilizantes con sabor a aceite de oliva" En este vídeo hemos preparado la reacción para poder amplificar el ADN por el método de la PCR. Más información en este enlace: http://www.compostandociencia.com/proyectos/piiisa2015-2016/ -

-

-

Practice 8: Ligation of a PCR Product in a Vector With Topoisomerase
-

" Tanging Ikaw " Jaker ✘ Double B ✘ Royxkie (PCR ENT.)
Rap | Music | Images PCR Entertainment 2016-2017 Recorded At: PCR Entertainment Title Of Song : Tanging Ikaw Mastered/Arranger : Joji Francisco Artist : Jaker, Double B & Royxkie Official Fanpage: https://www.facebook.com/pcrwreckordz For recording session info message this person : https://www.facebook.com/jojifrancisco03 Thanks a lot for your support! PCR Entertainment © 2016. All Rights Reserved. -

" Move On " Talim Ng Disiplina (PCR ENT.)
Rap | Music | Images PCR Entertainment 2016-2017 Recorded At: PCR Entertainment Title Of Song : Move On Mastered/Arranger : Joji Francisco Artist : Talim Ng Disiplina Official Fanpage: https://www.facebook.com/pcrwreckordz For recording session info message this person : https://www.facebook.com/jojifrancisco03 Thanks a lot for your support! PCR Entertainment © 2016. All Rights Reserved. -

" Buhat Ng Maging Tayo " Silent (PCR ENT.)
Rap | Music | Images PCR Entertainment 2016-2017 Recorded At: PCR Entertainment Title Of Song : Buhat Ng Maging Tayo Mastered/Arranger : Joji Francisco Artist : Silent Official Fanpage: https://www.facebook.com/pcrwreckordz For recording session info message this person : https://www.facebook.com/jojifrancisco03 Thanks a lot for your support! PCR Entertainment © 2016. All Rights Reserved.
SBV em PCR
- Order: Reorder
- Duration: 2:53
- Updated: 05 Apr 2016
- views: 6
Suporte basico de vida PCR ( Parada Cardio Respiratória) dos alunos do V termo de Enfermagem -Ajes
- Order: Reorder
- Duration: 5:15
- Updated: 05 Apr 2016
- views: 2
- published: 05 Apr 2016
- views: 2
PCR: College
- Order: Reorder
- Duration: 14:26
- Updated: 05 Apr 2016
- views: 5
Reportage PCR Vétérinaire Scanelis
- Order: Reorder
- Duration: 13:16
- Updated: 05 Apr 2016
- views: 12
- published: 05 Apr 2016
- views: 12
PCR van driver dies in accident
- Order: Reorder
- Duration: 0:49
- Updated: 05 Apr 2016
- views: 101
TEST DU CANON-PCR #1 |Clash royale
- Order: Reorder
- Duration: 5:06
- Updated: 04 Apr 2016
- views: 25
- published: 04 Apr 2016
- views: 25
BIGUP-PCR-2404
- Order: Reorder
- Duration: 0:33
- Updated: 04 Apr 2016
- views: 105
- published: 04 Apr 2016
- views: 105
The PCR Story
- Order: Reorder
- Duration: 5:52
- Updated: 04 Apr 2016
- views: 6
PCR | Delhi Aaj Tak | April 2, 2016 | 8:30 PM
- Order: Reorder
- Duration: 20:39
- Updated: 04 Apr 2016
- views: 38
PCR | Delhi Aaj Tak | April 3, 2016 | 8:30 PM
- Order: Reorder
- Duration: 43:41
- Updated: 04 Apr 2016
- views: 22
Casio PCR-T273 Electronic Cash Register
- Order: Reorder
- Duration: 2:19
- Updated: 04 Apr 2016
- views: 0
- published: 04 Apr 2016
- views: 0
forensic files s7e12 - PCR
- Order: Reorder
- Duration: 2:58
- Updated: 03 Apr 2016
- views: 20
- published: 03 Apr 2016
- views: 20
Portable and Efficient PCR Machine
- Order: Reorder
- Duration: 2:31
- Updated: 03 Apr 2016
- views: 3
- published: 03 Apr 2016
- views: 3
PCR - Burgh Boy (Prod. By Big Jus) [explicit]
- Order: Reorder
- Duration: 4:15
- Updated: 03 Apr 2016
- views: 2608
Project CARS. PCR:A 2016 Экзаменационная гонка - LIVE (03-04-2016)
- Order: Reorder
- Duration: 139:15
- Updated: 03 Apr 2016
- views: 167
- published: 03 Apr 2016
- views: 167
Polymerase Chain Reaction(PCR)-Understanding the steps through PCR machine
- Order: Reorder
- Duration: 4:07
- Updated: 03 Apr 2016
- views: 3
- published: 03 Apr 2016
- views: 3
Reacción de PCR
- Order: Reorder
- Duration: 4:31
- Updated: 03 Apr 2016
- views: 11
- published: 03 Apr 2016
- views: 11
PCR Simulation
- Order: Reorder
- Duration: 3:14
- Updated: 03 Apr 2016
- views: 9
Practice 8. PCR product ligation with topoisomerase
- Order: Reorder
- Duration: 10:11
- Updated: 03 Apr 2016
- views: 1
Practice 8: Ligation of a PCR Product in a Vector With Topoisomerase
- Order: Reorder
- Duration: 6:13
- Updated: 03 Apr 2016
- views: 0
- published: 03 Apr 2016
- views: 0
" Tanging Ikaw " Jaker ✘ Double B ✘ Royxkie (PCR ENT.)
- Order: Reorder
- Duration: 4:21
- Updated: 03 Apr 2016
- views: 23
- published: 03 Apr 2016
- views: 23
" Move On " Talim Ng Disiplina (PCR ENT.)
- Order: Reorder
- Duration: 4:36
- Updated: 03 Apr 2016
- views: 17
- published: 03 Apr 2016
- views: 17
" Buhat Ng Maging Tayo " Silent (PCR ENT.)
- Order: Reorder
- Duration: 4:42
- Updated: 03 Apr 2016
- views: 32
- published: 03 Apr 2016
- views: 32
-

線上課程-Real-Time PCR 原理線上課程
主講人 - 鄭婷之 儀器銷售專員, Life Technologies 台灣分公司 在這堂45分鐘的線上教學課程中,您將可以獲得儀器偵測原理、螢光化學原理介紹(包含TaqMan Probe & SYBR Green 作用機制),以及反應製備流程等相關知識。 -

DJ Private Ryan - PCR 2016 [2016 SOCA MIX DOWNLOAD]
The final mix to close off the Trinidad carnival 2016 season. Relive the memories of the parties and the road. Suffering from post carnival depression? Then this is the mix for you. 1 hour and 18 mins of the best. Enjoy!! Download: http://www.themixfeed.com/mixes/soca/private-ryan-presents-post-carnival-relief-the-anthems/ More mixes: http://www.theMixFeed.com -

Diagnóstico del virus del Papiloma Humano por PCR
Suscríbete gratis: http://www.actualizacionmedica.com Puedes obtener Horas Crédito de Educación Médica Continua (EMC) Únete a la Red de Actualización Medica y obtén muchos beneficios: Actualízate, Infórmate, e interrelaciona con tus colegas. Conferencista: Dr. Alberto Garcia Fecha: 29/11/1012 Lugar: Ciudad de Guatemala Evento: 54 Congreso Nacional de Medicina -

생명과학2- 생명공학: PCR 기술
http://cafe.daum.net/bioddong -

黒田裕樹の生物学講義〜分子生物学第11回『PCR』
分子生物学における発明の中でも、もっとも「発明!」という感じがする意味ではナンバーワンの技術革新は間違いなくPCR (ポリメラーゼ連鎖反応: polymerase chain reaction)だと思います。これは、変人科学者とも言われるドクター・マリス先生のアイデアで生まれたものですが(もちろんノーベル賞です)、私も初めて学んだ時には、感動させられたものでした。もし、過去に戻れたら、唱えてノーベル賞もらっちゃいたくなりますね。 では、この画期的な技術のメカニズム。とても簡単なものですので、ぜひマスターして下さいませ。 分子生物学 第11回『PCR』 1. 用途 2. 反応 3. Thermus aquaticus -

Deslocamento e atendimento a uma PCR!!!
Cada atendimento é uma história diferente! Neste atendimento passamos por várias dificuldades mas mesmo com todas elas nós lutamos até o fim pra salvar a vida alheia!! -

CZ 75 D Compact PCR Aluminum Framed Awesome!
Table top review for the CZ 75 D Compact PCR. In this video I give an introduction to all of the main functions of this gun and talk about some of my reasons for choosing it. Thanks for watching! If you enjoyed or have some constructive criticism please Like, Comment and Subscribe! Creative Commons Attribution: Kevin MacLeod - "Overriding Concern" -

-

P.C.R.Evo [2] - Evolução é acaso?
Será que a Evolução diz que tudo é ao acaso? Que somos fruto do acaso? Eu acho que não... Meu Twitter: @Pirulla25 Minha fanpage: https://www.facebook.com/CanalDoPirula Meu vídeo sobre Cladística: https://www.youtube.com/watch?v=SAoFkZczm2Y Sobre DNA (duplicação gênica, mutações, e sítios frágeis): https://en.wikipedia.org/wiki/Gene_duplication http://www.pnas.org/content/91/8/2950.short http://genetics.org/content/genetics/184/4/1077.full.pdf https://scholar.google.com.br/scholar?q=gene+duplication+frequency&hl;=en&as;_sdt=0&as;_vis=1&oi;=scholart&sa;=X&ved;=0ahUKEwjN2PvzyLjKAhXDgZAKHd-fBwMQgQMIJzAA http://mbe.oxfordjournals.org/content/22/1/135.full https://scholar.google.com.br/scholar?q=chromosome+fragile+portions&hl;=en&as;_sdt=0&as;_vis=1&oi;=scholart&sa;=X&ved;=0ahUKEwjdmpz5ybjKAhWMDZAKHXEPD2... -

-

Bài 1-PCR-Phần 1
Bài 1-PCR-Phần 1 -

Agarose Gel Electrophoresis, DNA Sequencing, PCR, Excerpt 2 | MIT 7.01SC Fundamentals of Biology
Agarose Gel Electrophoresis, DNA Sequencing, PCR, Excerpt 2 Instructor: Eric Lander View the complete course: http://ocw.mit.edu/7-01SCF11 License: Creative Commons BY-NC-SA More information at http://ocw.mit.edu/terms More courses at http://ocw.mit.edu -

Dj Private Ryan - Post Carnival Relief (PCR) 2015 - 2015 SOCA MIX
The final piece of the Carnival 2015 puzzle is here to cure your post carnival depression. Post Carnival Relief 2015 features all the road anthems for Trinidad Carnival 2015 all in 1 hour and 25 minutes. Includes all of your favourites artistes and road mixes as well. Enjoy your soca 2015 remedy!!! -

CZ 75 9mm Compact PCR. MIDDLEWEIGHT AWESOMENESS
test fire and review of the superb PCR from CZ. -

Discover Real-Time PCR for the Classroom
For more info, visit http://www.bio-rad.com/yt/1/qPCR. This Biotechnology Explorer™ webinar presents an overview of what real-time PCR is, what it is used for, and how it works. Also known as quantitative PCR (qPCR), this technique exploits the power of exponential DNA amplification by the polymerase chain reaction coupled with fluorescent detection of the products in real time, enabling researchers to detect and measure the initial number of copies of a target DNA sequence with high sensitivity and accuracy. Dr David Palmer, a Bay Area biotech entrepreneur and adjunct professor at Contra Costa College, explains the principles of qPCR with examples suitable for classroom instruction. Dr Palmer discusses the usefulness of this technology, citing real-world applications: measuring the expre... -

Exame PCR - Proteína C. Reativa - Parte 1 ( Prof.
CURSO AUXILIAR DE LABORATÓRIO -

Sichler NC 5725 PCR 3550UV Staubsaugerroboter Teil1-Test
Link Testbericht, Bewertung, Daten,Bezug: http://www.akku-und-roboter-staubsauger.de/staubsaugerroboter-sichler-nc-5725-pcr-3550uv-im-test Bezugsquellen findet ihr hier. http://www.akku-und-roboter-staubsauger.de/tools/?search_az=Sichler,3550 Ein preiswerter Roboterstaubsauger muss nicht immer schlecht sein, das hat mein neuer Test von dem Staubsaugerroboter Sichler NC-5725 gezeigt. Das Modell wird auch unter dem Namen Sichler PCR-3550UV angeboten. Im Test Schnitt der Roboter bislang als bester in der Preisklasse bis 200 Euro ab, siehe Testbericht! Schaut euch auch Teil2 des Videos mit den Härtetests an. Video findet ihr auch im Testbericht oder bei Youtube. Weitere Staubsaugerroboter und Akkustaubsauger im Test und Vergleich auf unserer Seite http://www.akku-und-roboter-staubsauger.de -

An introduction to real-time PCR and examples of its applications
Real-time PCR is already well established in many fields, including food safety and quality testing and environmental assessment. It enables the rapid and sensitive detection of specific nucleic acids.
線上課程-Real-Time PCR 原理線上課程
- Order: Reorder
- Duration: 45:52
- Updated: 14 Jan 2013
- views: 4711
- published: 14 Jan 2013
- views: 4711
DJ Private Ryan - PCR 2016 [2016 SOCA MIX DOWNLOAD]
- Order: Reorder
- Duration: 80:32
- Updated: 12 Mar 2016
- views: 1991
- published: 12 Mar 2016
- views: 1991
Diagnóstico del virus del Papiloma Humano por PCR
- Order: Reorder
- Duration: 25:15
- Updated: 05 Jan 2013
- views: 19120
- published: 05 Jan 2013
- views: 19120
생명과학2- 생명공학: PCR 기술
- Order: Reorder
- Duration: 40:26
- Updated: 10 Nov 2014
- views: 1533
黒田裕樹の生物学講義〜分子生物学第11回『PCR』
- Order: Reorder
- Duration: 55:12
- Updated: 01 Nov 2011
- views: 10911
- published: 01 Nov 2011
- views: 10911
Deslocamento e atendimento a uma PCR!!!
- Order: Reorder
- Duration: 30:30
- Updated: 21 Jul 2015
- views: 16404
- published: 21 Jul 2015
- views: 16404
CZ 75 D Compact PCR Aluminum Framed Awesome!
- Order: Reorder
- Duration: 25:19
- Updated: 01 Nov 2015
- views: 9200
- published: 01 Nov 2015
- views: 9200
PCR
- Order: Reorder
- Duration: 25:46
- Updated: 24 Mar 2013
- views: 4774
P.C.R.Evo [2] - Evolução é acaso?
- Order: Reorder
- Duration: 29:12
- Updated: 20 Jan 2016
- views: 109772
- published: 20 Jan 2016
- views: 109772
Обзор катушки Preston PCR 5000
- Order: Reorder
- Duration: 20:15
- Updated: 17 Sep 2015
- views: 2782
Bài 1-PCR-Phần 1
- Order: Reorder
- Duration: 25:52
- Updated: 20 Jul 2015
- views: 1445
- published: 20 Jul 2015
- views: 1445
Agarose Gel Electrophoresis, DNA Sequencing, PCR, Excerpt 2 | MIT 7.01SC Fundamentals of Biology
- Order: Reorder
- Duration: 42:27
- Updated: 09 May 2012
- views: 55399
- published: 09 May 2012
- views: 55399
Dj Private Ryan - Post Carnival Relief (PCR) 2015 - 2015 SOCA MIX
- Order: Reorder
- Duration: 86:08
- Updated: 04 Mar 2015
- views: 598937
- published: 04 Mar 2015
- views: 598937
CZ 75 9mm Compact PCR. MIDDLEWEIGHT AWESOMENESS
- Order: Reorder
- Duration: 28:19
- Updated: 12 Aug 2014
- views: 25431
Discover Real-Time PCR for the Classroom
- Order: Reorder
- Duration: 61:00
- Updated: 29 Jan 2013
- views: 18925
- published: 29 Jan 2013
- views: 18925
Exame PCR - Proteína C. Reativa - Parte 1 ( Prof.
- Order: Reorder
- Duration: 20:01
- Updated: 12 Jun 2014
- views: 8150
- published: 12 Jun 2014
- views: 8150
Sichler NC 5725 PCR 3550UV Staubsaugerroboter Teil1-Test
- Order: Reorder
- Duration: 26:05
- Updated: 18 Jan 2016
- views: 1083
- published: 18 Jan 2016
- views: 1083
An introduction to real-time PCR and examples of its applications
- Order: Reorder
- Duration: 47:59
- Updated: 12 Jun 2014
- views: 4874
- published: 12 Jun 2014
- views: 4874
- Playlist
- Chat
- Playlist
- Chat

PCR (Polymerase Chain Reaction)
- Report rights infringement
- published: 20 Apr 2014
- views: 470310

Polymerase chain reaction (PCR)
- Report rights infringement
- published: 25 Mar 2015
- views: 44258

Polymerase Chain Reaction (PCR)
- Report rights infringement
- published: 20 Jan 2015
- views: 21303

Polymerase Chain Reaction (PCR)
- Report rights infringement
- published: 22 Mar 2010
- views: 483263

Polymerase Chain Reaction (PCR)
- Report rights infringement
- published: 16 Apr 2009
- views: 444274

Die PCR-Methode einfach erklärt
- Report rights infringement
- published: 08 Dec 2014
- views: 167402

Polymerase Chain Reaction (PCR) | MIT 7.01SC Fundamentals of Biology
- Report rights infringement
- published: 09 May 2012
- views: 141328

PCR - Polymerase Chain Reaction - Simple Animated Tutorial
- Report rights infringement
- published: 02 Jun 2012
- views: 214715

How to Set Up the PCR Reaction
- Report rights infringement
- published: 28 Feb 2013
- views: 74134

PCR (polymerase chain reaction) in detail
- Report rights infringement
- published: 27 Oct 2013
- views: 62960

The PCR Song
- Report rights infringement
- published: 08 Jan 2008
- views: 1641093

Aula de PCR e eletroforese
- Report rights infringement
- published: 16 Oct 2014
- views: 27246

PCR for A-level
- Report rights infringement
- published: 16 May 2012
- views: 45032
- Playlist
- Chat


Suporte basico de vida PCR ( Parada Cardio Respiratória) dos alunos do V termo de Enfermagem -Ajes
- Report rights infringement
- published: 05 Apr 2016
- views: 2

PCR: College
- Report rights infringement
- published: 05 Apr 2016
- views: 5

Reportage PCR Vétérinaire Scanelis
- Report rights infringement
- published: 05 Apr 2016
- views: 12

PCR van driver dies in accident
- Report rights infringement
- published: 05 Apr 2016
- views: 101

TEST DU CANON-PCR #1 |Clash royale
- Report rights infringement
- published: 04 Apr 2016
- views: 25

BIGUP-PCR-2404
- Report rights infringement
- published: 04 Apr 2016
- views: 105

The PCR Story
- Report rights infringement
- published: 04 Apr 2016
- views: 6

PCR | Delhi Aaj Tak | April 2, 2016 | 8:30 PM
- Report rights infringement
- published: 04 Apr 2016
- views: 38

PCR | Delhi Aaj Tak | April 3, 2016 | 8:30 PM
- Report rights infringement
- published: 04 Apr 2016
- views: 22

Casio PCR-T273 Electronic Cash Register
- Report rights infringement
- published: 04 Apr 2016
- views: 0

forensic files s7e12 - PCR
- Report rights infringement
- published: 03 Apr 2016
- views: 20

Portable and Efficient PCR Machine
- Report rights infringement
- published: 03 Apr 2016
- views: 3

PCR - Burgh Boy (Prod. By Big Jus) [explicit]
- Report rights infringement
- published: 03 Apr 2016
- views: 2608
- Playlist
- Chat

線上課程-Real-Time PCR 原理線上課程
- Report rights infringement
- published: 14 Jan 2013
- views: 4711

DJ Private Ryan - PCR 2016 [2016 SOCA MIX DOWNLOAD]
- Report rights infringement
- published: 12 Mar 2016
- views: 1991

Diagnóstico del virus del Papiloma Humano por PCR
- Report rights infringement
- published: 05 Jan 2013
- views: 19120

생명과학2- 생명공학: PCR 기술
- Report rights infringement
- published: 10 Nov 2014
- views: 1533

黒田裕樹の生物学講義〜分子生物学第11回『PCR』
- Report rights infringement
- published: 01 Nov 2011
- views: 10911

Deslocamento e atendimento a uma PCR!!!
- Report rights infringement
- published: 21 Jul 2015
- views: 16404

CZ 75 D Compact PCR Aluminum Framed Awesome!
- Report rights infringement
- published: 01 Nov 2015
- views: 9200


P.C.R.Evo [2] - Evolução é acaso?
- Report rights infringement
- published: 20 Jan 2016
- views: 109772

Обзор катушки Preston PCR 5000
- Report rights infringement
- published: 17 Sep 2015
- views: 2782

Bài 1-PCR-Phần 1
- Report rights infringement
- published: 20 Jul 2015
- views: 1445

Agarose Gel Electrophoresis, DNA Sequencing, PCR, Excerpt 2 | MIT 7.01SC Fundamentals of Biology
- Report rights infringement
- published: 09 May 2012
- views: 55399

Dj Private Ryan - Post Carnival Relief (PCR) 2015 - 2015 SOCA MIX
- Report rights infringement
- published: 04 Mar 2015
- views: 598937

CZ 75 9mm Compact PCR. MIDDLEWEIGHT AWESOMENESS
- Report rights infringement
- published: 12 Aug 2014
- views: 25431
-
Lyrics list:text lyricsplay full screenplay karaoke
The Panama papers could hand Bernie Sanders the keys to the White House
Edit The Independent 05 Apr 2016Apple is close to selling its billionth iPhone
Edit Topix 05 Apr 2016Who Is Chris Gould? Get to Know Mimi Faust’s New Lover
Edit VH1 05 Apr 2016Panama Papers: UK PM under pressure over family wealth
Edit Dawn 05 Apr 2016Can We Take a Selfie Before You Blow Me Up?
Edit WorldNews.com 05 Apr 2016Assistant sub inspector held for abetting Dhruv's suicide
Edit The Times of India 05 Apr 2016Biodist y Seegene crean empresa conjunta en México
Edit PR Newswire 05 Apr 2016Global Molecular Diagnostics Technologies, Markets and Companies Study 2016-2025 - 342 Companies Profiled with Tabulation ...
Edit Business Wire 05 Apr 2016IAEA Trains Experts to Use Diagnostic Tools for Quick Zika Detection (IAEA - International Atomic Energy Agency)
Edit Public Technologies 05 Apr 2016Traffic restrictions imposed around LB stadium
Edit The Times of India 05 Apr 2016UN agency to train dozens of experts on nuclear-related techniques to identify Zika
Edit United Nations 05 Apr 2016UN agency to train dozens of experts on nuclear-related techniques to identify Zika (UN - United Nations)
Edit Public Technologies 05 Apr 2016Drone spotted in outer Delhi, this time it's CRPF's
Edit The Times of India 05 Apr 2016‘I stopped to help hearing girl’s cries’
Edit The Times of India 05 Apr 2016CombiMatrix Launches Preimplantation Genetic Diagnostic Testing for Single Gene Disorders and Chromosomal Translocations
Edit Stockhouse 05 Apr 2016CombiMatrix Launches Preimplantation Genetic Diagnostic Testing for Single Gene Disorders and Chromosomal Translocations (CombiMatrix Corporation)
Edit Public Technologies 05 Apr 2016Transcript for CDC Telebriefing: Zika Summit Press Conference - Transcript (CDC - Centers for Disease Control and Prevention)
Edit Public Technologies 05 Apr 2016ASI held for abetting Dhruv's suicide
Edit The Times of India 04 Apr 2016- 1
- 2
- 3
- 4
- 5
- Next page »








