- published: 18 Jul 2014
- views: 80
- published: 16 Jul 2012
- views: 256
- published: 23 May 2014
- views: 339
- published: 10 Feb 2014
- views: 481
- published: 11 Dec 2014
- views: 163
- published: 23 Dec 2014
- views: 191
- published: 21 Apr 2015
- views: 206
- published: 28 Mar 2013
- views: 125
- published: 12 Sep 2013
- views: 337
- published: 12 Dec 2013
- views: 138
- published: 10 Dec 2014
- views: 96
- Loading...

-
 1:59
1:59Dr. Hurvitz on Patient Outcomes and PIK3CA Mutations
Dr. Hurvitz on Patient Outcomes and PIK3CA MutationsDr. Hurvitz on Patient Outcomes and PIK3CA Mutations
Sara Hurvitz, MD, medical oncologist, UCLA Medical Center, discusses patient outcomes associated with PIK3CA mutations. For more resources and information regarding anticancer targeted therapies: http://targetedonc.com/ -
 8:34
8:34PIK3CA Mutational Status and Survival in Locally Advanced Cervical Cancer Patients
PIK3CA Mutational Status and Survival in Locally Advanced Cervical Cancer PatientsPIK3CA Mutational Status and Survival in Locally Advanced Cervical Cancer Patients
John B McIntyre, PhD, Peter S Craighead, MD, Tien Phan, MD, Prafull Ghatage, MD, Anthony M Magliocco, MD, Corinne M Doll, MD. Department of Oncology, University of Calgary -
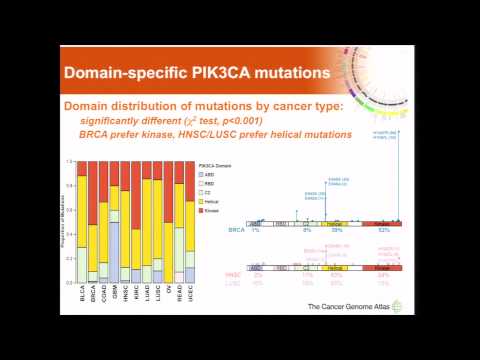 15:01
15:01Domain-specific PIK3CA mutations affect different pathway activities... - Christopher Benz
Domain-specific PIK3CA mutations affect different pathway activities... - Christopher BenzDomain-specific PIK3CA mutations affect different pathway activities... - Christopher Benz
May 12, 2014 - The Cancer Genome Atlas 3rd Annual Scientific Symposium More: http://www.genome.gov/27557040 -
 4:20
4:20PIK3CA Mutations in HER2-Positive Breast Cancer
PIK3CA Mutations in HER2-Positive Breast CancerPIK3CA Mutations in HER2-Positive Breast Cancer
Panelists discuss the possible deleterious impact of PIK3CA mutations in patients with HER2-positive breast cancer receiving treatment with HER2-targeted therapies. For more from this discussion, visit http://www.onclive.com/peer-exchange/breast-cancer-therapies -
 4:19
4:19FISH analysis of HSR type PIK3CA amplification in endometrial cancer
FISH analysis of HSR type PIK3CA amplification in endometrial cancerFISH analysis of HSR type PIK3CA amplification in endometrial cancer
Analysis of a HSR type phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha gene (PIK3CA) amplification detected by fluorescent in situ hybridization (FISH) in endometrial cancer: The PIK3CA FISH probe (Abnova, Taipei City, Taiwan) is labeled with a “Texas Red®” fluorochrophor. Gene signals appear as red pinhead shaped dots. For copy number reference a chromosome enumeration probe for centromere of Chromosome 3 (CEP3) is labeled with a “FITC” fluorochphor. CEP3 signals appear as green pinhead shaped dots. Nuclei are counterstained with 4’,6-diamino-2-phenylindole (DAPI). Nuclear chromatin DNA is appearing in blue color. Video clip starts with microscope fluorescence filter setting for DAPI staining: Chromatin stained nuclei are checked. Sufficient integer and separable nuclei are selected to determine gene and centromere copy number. Full z-axis is taken into account. It has also to be taken into account, that a complete integrity of nuclei can not be expected or guaranteed for most nuclei analyzed, since a risk of truncation of nuclei due to tissue slide preparation, is always given in 4 µm tissue sections. Filter change to “Texas Red®”: Distribution of FISH signals over cell nuclei of tissues can mostly be estimated for a first sight screening in the fluorescence filter spectrum of the gene probe fluorochrophor. Signals in full z-axis are taken into account. PIK3CA signals in this tumor occur predominantly in a pattern of clustered signal distribution of HSR type (homogeneously staining region). Clusters of two, three up six or seven signals signals nearby to each other, but also dislocated additional gene signals occur. Based on previously observed DAPI staining and visible gene signals, nuclei are selected for counting. Number of gene signals is determined. The three dimensional constitution of the nucleus is taken into account by a variable adjustment of optical z-axis layer. First nucleus (first white arrow) shows a tiny signal cluster of four signals tight nearby each other at one chromosomal allelic location, but only one signal on probably the other chromosome. Second selected nucleus (second arrow) shows about six or seven gene signals, partly in a “cluster” pattern of two or three signals nearby each other. Filter change to “FITC” (green): Signals of the centromere reference, the chromosome enumeration probe (CEP), are checked for signal quality. Signals in full z-axis are taken into account. CEP3 signals of the centromere reference are counted. Signals in full z-axis are taken into account. The first and second nucleus both show show signals for two centromere loci indicating presence of two chromosomes. Filter change to “Texas Red®”: Tissue is screened for additional nuclei suited for gene copy number determination. The third nucleus (third white arrow) shows a signal cluster of about five or six signals tight nearby each other and four additional distributed signals on other chromosomal other extra-chromosomal locations. The fourth selected nucleus (fourth arrow) shows four or even six gene signals, although the three or four gene signals marked by the arrow might be allelic gene copy signals of four chromatids as part of two chromosomes. Nevertheless, the another gene signals of the same nucleus appear as single point shape signals, indicating not to be copied on another chromatid. However, the nucleus shows at least four gene copies. Filter change to “FITC” (green): Signals of the centromere reference, the chromosome enumeration probe (CEP), are checked for signal quality. Signals in full z-axis are taken into account. CEP3 signals of the centromere reference are counted. Signals in full z-axis are taken into account. The third and fourth nucleus both show show signals for two centromere loci indicating presence of two chromosomes. Filter change to DAPI (blue): Chromatin stained nuclei are checked for integrity and separability. This video presentation is the intellectual property of the University of Bergen, Norway and is published for scientific research purpose in line with regarding ethical laws. Correspondence: frederik.holst@k2.uib.no -
 1:50
1:50Randox KRAS BRAF PIK3CA* Array - Molecular Testing
Randox KRAS BRAF PIK3CA* Array - Molecular TestingRandox KRAS BRAF PIK3CA* Array - Molecular Testing
The KRAS, BRAF, PIK3CA* Array is designed for the rapid qualitative simultaneous detection of point mutations within the KRAS, BRAF and PIK3CA* genes from fresh/frozen and formalin fixed paraffin embedded (FFPE) tissue DNA for predictive and prognostic mutation profiling. *PIK3CA for research use only -
 9:28
9:28Are the benefits of aspirin in colorectal cancer limited to PIK3CA mutated cancers?
Are the benefits of aspirin in colorectal cancer limited to PIK3CA mutated cancers?Are the benefits of aspirin in colorectal cancer limited to PIK3CA mutated cancers?
Alberto Sobrero provides a critical analysis of evidence currently available that mutations in the PIK3CA gene in colorectal cancer might identify patients that receive greater benefit from aspirin in the adjuvant setting. http://www.esmo.org Video produced by the European Society for Medical Oncology (ESMO) -
 1:02
1:02Dr. Baselga Discusses PIK3CA Inhibitors in Breast Cancer
Dr. Baselga Discusses PIK3CA Inhibitors in Breast CancerDr. Baselga Discusses PIK3CA Inhibitors in Breast Cancer
José Baselga, MD, PhD, Physician-in-Chief, Memorial Sloan-Kettering Cancer Center, discusses treatment with PIK3CA inhibitors in patients with breast cancer. For more information and resources on breast cancer: http://www.onclive.com/specialty/breast-cancer -
 4:44
4:44The PI3K/AKT signalling pathway
The PI3K/AKT signalling pathwayThe PI3K/AKT signalling pathway
-
 9:18
9:18Role of TGF-beta signaling in PIK3CA-driven Head and Neck Cancer Invasion and Metastasis
Role of TGF-beta signaling in PIK3CA-driven Head and Neck Cancer Invasion and MetastasisRole of TGF-beta signaling in PIK3CA-driven Head and Neck Cancer Invasion and Metastasis
Sophia Bornstein, MD, PhD, Jingping Shen, MD, PhD, Frank Hall, BS, Sherif Said, MD, PhD, Xiao-Jing Wang, MD, PhD, Neil Gross, MD, John Song, MD, Natalie Serkova, PhD, Shi-Long Lu, MD, PhD. Oregon Health & Science University, Portland, OR (Departments of Otolaryngology, Radiation Medicine); University of Colorado Anschutz Medical Campus, Aurora, CO (Departments of Otolaryngology, Pathology, Anesthesiology) -
 10:59
10:59Sibylle Loibl: PIK3CA mutation predicts resistance to anti HER2/chemotherapy
Sibylle Loibl: PIK3CA mutation predicts resistance to anti HER2/chemotherapySibylle Loibl: PIK3CA mutation predicts resistance to anti HER2/chemotherapy
Thursday, Dec. 12 at 7:30 a.m. CT, a 3rd press conference hosted by Carlos L. Arteaga, M.D., president-elect of the AACR and associate director for translational/clinical research and director of the Breast Cancer Program at Vanderbilt-Ingram Comprehensive Cancer Center, will feature the following research: Sibylle Loibl: PIK3CA mutation predicts resistance to anti HER2/chemotherapy in primary HER2 positive/hormone-receptor-positive breast cancer Prospective analysis of 737 participants of the GeparSixto and GeparQuinto studies More info: http://oncoletter.ch -
 3:48
3:48FISH analysis of a typical PIK3CA amplification in endometrial cancer.
FISH analysis of a typical PIK3CA amplification in endometrial cancer.FISH analysis of a typical PIK3CA amplification in endometrial cancer.
Analysis of a typical phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha gene (PIK3CA) amplification detected by fluorescent in situ hybridization (FISH) in endometrial cancer: The PIK3CA FISH probe (Abnova, Taipei City, Taiwan) is labeled with a “Texas Red®” fluorochrophor. Gene signals appear as red pinhead shaped dots. For copy number reference a chromosome enumeration probe for centromere of Chromosome 3 (CEP3) is labeled with a “FITC” fluorochphor. CEP3 signals appear as green pinhead shaped dots. Nuclei are counterstained with 4’,6-diamino-2-phenylindole (DAPI). Nuclear chromatin DNA is appearing in blue color. Video clip starts with microscope fluorescence filter setting for fluorochrophor “Texas Red®”: Distribution of FISH signals over cell nuclei of tissues can mostly be estimated for a first sight screening in the fluorescence filter spectrum of the gene probe fluorochrophor. Signals in full z-axis are taken into account. Although most PIK3CA signals in this tumor occur in a pattern with large distance of gene signal distribution, small “clusters” of two or three signals nearby to each other occur also in this specimen (marked by the first three white arrows). Size and shape of nuclei can be estimated by their weak red “background” fluorescence and two non overlapping nuclei are selected for counting. Number of gene signals is determined. The three dimensional constitution of the nucleus is taken into account by a variable adjustment of optical z-axis layer. The first nucleus (fourth white arrow) shows four and the second selected nucleus (fifth arrow) shows five gene signals. One of these five signals of the second nucleus is clearly smaller than the other four. Filter change to “FITC” (green): Signals of the centromere reference, the chromosome enumeration probe (CEP), are checked for signal quality. Signals in full z-axis are taken into account. CEP3 signals of the centromere reference are counted. Signals in full z-axis are taken into account. The first nucleus shows two and the second selected nucleus shows three centromere signals. Filter change to “Texas Red®”: Number of gene signals and representativeness of tumor area is checked again. Filter change to DAPI (blue): Chromatin stained nuclei are checked. Sufficient integer and separable nuclei are selected to determine gene and centromere copy number. Full z-axis is taken into account. It has also to be taken into account, that a complete integrity of nuclei can not be expected or guaranteed for most nuclei analyzed, since a risk of truncation of nuclei due to tissue slide preparation, is always given in 4 µm tissue sections. This video presentation is the intellectual property of the University of Bergen, Norway and is published for scientific research purpose in line with regarding ethical laws. Correspondence: frederik.holst@k2.uib.no -
 3:08
3:08Aspirin Use, Tumor PIK3CA Mutation, and Colorectal Cancer Su
Aspirin Use, Tumor PIK3CA Mutation, and Colorectal Cancer SuAspirin Use, Tumor PIK3CA Mutation, and Colorectal Cancer Su
-
 6:05
6:05Whitney Henry | Harvard Horizons Symposium
Whitney Henry | Harvard Horizons SymposiumWhitney Henry | Harvard Horizons Symposium
April 2014: Harvard Horizons Symposium, featuring Whitney Henry, a PhD candidate in Biological and Biomedical Sciences, on "Understanding the Chemotherapeutic Benefit of Aspirin in Mutant PIK3CA breast cancer" Harvard Horizons, an initiative at the Graduate School of Arts and Sciences to celebrate the ideas and innovations of PhD students at Harvard, selected 8 finalists as 2014 Horizon Scholars. Their talks were delivered at the Harvard Horizons Symposium in Sanders Theatre on April 22, 2014. Read about Harvard Horizons and the 2014 Horizon Scholars: http://www.gsas.harvard.edu/harvardhorizons
-

Dr. Hurvitz on Patient Outcomes and PIK3CA Mutations
Sara Hurvitz, MD, medical oncologist, UCLA Medical Center, discusses patient outcomes associated with PIK3CA mutations. For more resources and information regarding anticancer targeted therapies: http://targetedonc.com/ -

PIK3CA Mutational Status and Survival in Locally Advanced Cervical Cancer Patients
John B McIntyre, PhD, Peter S Craighead, MD, Tien Phan, MD, Prafull Ghatage, MD, Anthony M Magliocco, MD, Corinne M Doll, MD. Department of Oncology, University of Calgary -

Domain-specific PIK3CA mutations affect different pathway activities... - Christopher Benz
May 12, 2014 - The Cancer Genome Atlas 3rd Annual Scientific Symposium More: http://www.genome.gov/27557040 -

PIK3CA Mutations in HER2-Positive Breast Cancer
Panelists discuss the possible deleterious impact of PIK3CA mutations in patients with HER2-positive breast cancer receiving treatment with HER2-targeted therapies. For more from this discussion, visit http://www.onclive.com/peer-exchange/breast-cancer-therapies -

FISH analysis of HSR type PIK3CA amplification in endometrial cancer
Analysis of a HSR type phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha gene (PIK3CA) amplification detected by fluorescent in situ hybridization (FISH) in endometrial cancer: The PIK3CA FISH probe (Abnova, Taipei City, Taiwan) is labeled with a “Texas Red®” fluorochrophor. Gene signals appear as red pinhead shaped dots. For copy number reference a chromosome enumeration probe for centromere of Chromosome 3 (CEP3) is labeled with a “FITC” fluorochphor. CEP3 signals appear as green pinhead shaped dots. Nuclei are counterstained with 4’,6-diamino-2-phenylindole (DAPI). Nuclear chromatin DNA is appearing in blue color. Video clip starts with microscope fluorescence filter setting for DAPI staining: Chromatin stained nuclei are checked. Sufficient integer and separa... -

Randox KRAS BRAF PIK3CA* Array - Molecular Testing
The KRAS, BRAF, PIK3CA* Array is designed for the rapid qualitative simultaneous detection of point mutations within the KRAS, BRAF and PIK3CA* genes from fresh/frozen and formalin fixed paraffin embedded (FFPE) tissue DNA for predictive and prognostic mutation profiling. *PIK3CA for research use only -

Are the benefits of aspirin in colorectal cancer limited to PIK3CA mutated cancers?
Alberto Sobrero provides a critical analysis of evidence currently available that mutations in the PIK3CA gene in colorectal cancer might identify patients that receive greater benefit from aspirin in the adjuvant setting. http://www.esmo.org Video produced by the European Society for Medical Oncology (ESMO) -

Dr. Baselga Discusses PIK3CA Inhibitors in Breast Cancer
José Baselga, MD, PhD, Physician-in-Chief, Memorial Sloan-Kettering Cancer Center, discusses treatment with PIK3CA inhibitors in patients with breast cancer. For more information and resources on breast cancer: http://www.onclive.com/specialty/breast-cancer -

The PI3K/AKT signalling pathway
-

Role of TGF-beta signaling in PIK3CA-driven Head and Neck Cancer Invasion and Metastasis
Sophia Bornstein, MD, PhD, Jingping Shen, MD, PhD, Frank Hall, BS, Sherif Said, MD, PhD, Xiao-Jing Wang, MD, PhD, Neil Gross, MD, John Song, MD, Natalie Serkova, PhD, Shi-Long Lu, MD, PhD. Oregon Health & Science University, Portland, OR (Departments of Otolaryngology, Radiation Medicine); University of Colorado Anschutz Medical Campus, Aurora, CO (Departments of Otolaryngology, Pathology, Anesthesiology) -

Sibylle Loibl: PIK3CA mutation predicts resistance to anti HER2/chemotherapy
Thursday, Dec. 12 at 7:30 a.m. CT, a 3rd press conference hosted by Carlos L. Arteaga, M.D., president-elect of the AACR and associate director for translational/clinical research and director of the Breast Cancer Program at Vanderbilt-Ingram Comprehensive Cancer Center, will feature the following research: Sibylle Loibl: PIK3CA mutation predicts resistance to anti HER2/chemotherapy in primary HER2 positive/hormone-receptor-positive breast cancer Prospective analysis of 737 participants of the GeparSixto and GeparQuinto studies More info: http://oncoletter.ch -

FISH analysis of a typical PIK3CA amplification in endometrial cancer.
Analysis of a typical phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha gene (PIK3CA) amplification detected by fluorescent in situ hybridization (FISH) in endometrial cancer: The PIK3CA FISH probe (Abnova, Taipei City, Taiwan) is labeled with a “Texas Red®” fluorochrophor. Gene signals appear as red pinhead shaped dots. For copy number reference a chromosome enumeration probe for centromere of Chromosome 3 (CEP3) is labeled with a “FITC” fluorochphor. CEP3 signals appear as green pinhead shaped dots. Nuclei are counterstained with 4’,6-diamino-2-phenylindole (DAPI). Nuclear chromatin DNA is appearing in blue color. Video clip starts with microscope fluorescence filter setting for fluorochrophor “Texas Red®”: Distribution of FISH signals over cell nuclei of tissue... -

Aspirin Use, Tumor PIK3CA Mutation, and Colorectal Cancer Su
-

Whitney Henry | Harvard Horizons Symposium
April 2014: Harvard Horizons Symposium, featuring Whitney Henry, a PhD candidate in Biological and Biomedical Sciences, on "Understanding the Chemotherapeutic Benefit of Aspirin in Mutant PIK3CA breast cancer" Harvard Horizons, an initiative at the Graduate School of Arts and Sciences to celebrate the ideas and innovations of PhD students at Harvard, selected 8 finalists as 2014 Horizon Scholars. Their talks were delivered at the Harvard Horizons Symposium in Sanders Theatre on April 22, 2014. Read about Harvard Horizons and the 2014 Horizon Scholars: http://www.gsas.harvard.edu/harvardhorizons -

Mario Campone
SABCS2015 Press Briefing 3: Phase III, BELLE-2 Trial Meets Primary Endpoint of Progression-free Survival: Abstract S6-01[nbsp]José Baselga, Seock-Ah Im, Hiroji Iwata, et al. Presented by Mario Campone PIK3CA status in circulating tumor DNA (ctDNA) predicts efficacy of buparlisib (BUP) plus fulvestrant (FULV) in postmenopausal women with endocrine-resistant HR+/HER2– advanced breast cancer (BC): First results from the randomized, phase III BELLE-2 trial. -

Rapid Diagnostic Testing in Molecular Pathology
For a number of cancersubtypes it is crucial to determine if a mutation is present in genes like KRAS, EGFR, BRAF and PIK3CA. The testresults will be consequently used to predict the response to therapy. We demonstrate the workflow of this process in the Molecular Tumour Diagnostics group in the department of Pathology at the Leiden University Medical Center in the Netherlands. Citation: Rapid KRAS, EGFR, BRAF and PIK3CA Mutation Analysis of Fine Needle Aspirates from Non-Small-Cell Lung Cancer Using Allele-Specific qPCR. van Eijk R, Licht J, Schrumpf M, Talebian Yazdi M, Ruano D, Giusi I. Forte, Petra M. Nederlof, Maud Veselic, Klaus F. Rabe, Jouke T. Annema, Vincent Smit, Hans Morreau, Tom van Wezel. (2011) PLoS ONE 6(3): e17791. doi:10.1371/journal.pone.0017791 -

Liquid Biopsy Clinical Research – Tracking Mutations in cfDNA and CTCs
Advances in technology such as automated end-to-end solutions are allowing clinical researchers to study tough liquid biopsy samples. Leaders in the field have designed next- generation sequencing (NGS) assays that target critical questions in cancer research and tumor biology. We recently caught up with Luca Quagliata, MD, of University Hospital, Basel, Switzerland. We discussed tracking mutations in metastatic prostate cancer using cell-free DNA (cfDNA) and circulating tumor cells (CTCs). Part of our conversation included details of the protocol he’s using to conduct NGS analysis on 20–30 ng of DNA he obtains from 10 mL starting blood sample using the LiquidBiopsy™ Platform. Dr. Quagliata shared that many of his European colleagues are moving to targeted sequencing using multi-target a... -

AACR 2009: Genetic influences in response to EGFR for metastatic colorectal cancer
Frederica Di Nicolantonio, Pharm.D., Ph.D., research fellow, Laboratory of Molecular Genetics, The Institute for Cancer Research and Treatment, University of Turin Medical School, speaking at AACR 2009, Denver: BRAF, PIK3CA and KRAS Mutations and Loss of PTEN Expression Impair Response to EGFR-Targeted Therapies in Metastatic Colorectal Cancer -

Dr. Kris Discusses Available Targeted Therapies
Lead author Mark G. Kris, MD, chief of the Thoracic Oncology Service at Memorial Sloan-Kettering Cancer Center, describes the targeted agents that are available for some of the driver mutations examined in his multicenter study. The trial focused on identifying KRAS, EGFR, HER2, BRAF, PIK3CA, AKT1, MEK1, and NRAS mutations using standard multiplexed assays and FISH for EML4-ALK rearrangements and MET amplifications. Kris explains that erlotinib, which targets the EGFR mutation, is effective in nearly 20% of lung cancer patients. He anticipates that crizotinib will soon be approved which will be effective in approximately 7% patients. Other drugs, such as trastuzumab for HER2 positive non-small cell lung cancer, are also being developed or investigated for the remaining mutations examined ... -

Rare Mutation Detection w/ Digital PCR - ASHG 2014
Learn more at http://www.lifetechnologies.com/us/en/home/life-science/pcr/digital-pcr/rare-mutation-analysis.html.html Among several technologies on display at the American Society for Human Genetics annual conference, digital PCR certainly has garnered some buzz over the past couple of years. The small but mighty QuantStudio 3D digital PCR system made its public debut two years ago in San Francisco, and it has some great enhancements to boot this year in San Diego – specifically around rare mutation analysis. Product manager Dr. Iain Russell spoke with us about the new QuantStudio® 3D rare mutation analysis solution. The solution includes a subset of wet lab–validated TaqMan SNP Genotyping Assays to detect and quantify the most common cancer-related mutations (e.g., EGFR, BRAF, KRAS, PI... -

5 Women Who You Won’t Believe Actually Exist
5 Women Who You Won’t Believe Actually Exist :- 1) Julia Gunse developed a skin condition called porphyria. When medical Treatment offered limited help Julia slowly covered her body and face with Tattoos. Today, over 95% of her body is covered in tattoos with a journey of five years to create at the cost of over $80,000.00. Today she is listed in the Guinness World Records as most tattooed female in the world. 2) Mandy Sellars from birth battled with legs that would not stop growing. Her condition is known as Proteus syndrome. Doctors discovered a PIK3CA gene mutation in Mandy who is the only woman in the world to have it. Her legs are three times larger than normal legs with each weighing about 50 pounds. After a series of complications Mandy had her left leg amputated. What is more am... -

Buparlisib helps overcome endocrine resistance in metastatic breast cancer: BELLE-2 trial results
Dr Campone talks to ecancertv at SABCS 2015 about the first results from the randomised, phase III BELLE-2 trial that assessed the the efficacy and safety of combining the PI3K inhibitor buparlisib with the endocrine therapy fulvestrant in postmenopausal women with endocrine resistant, hormone receptor (HR)-positive, HER2-negative metastatic breast cancer. Activation of the PI3K pathway is a hallmark of resistance to endocrine therapy, Dr Campone explains. Early data suggest that using a PI3K inhibitor may be able to help reverse endocrine resistance and so the BELLE-2 trial was designed to look at this question further. More than 1,140 women who were refractory to aromatase inhibitor therapy were enrolled into the study and stratified based on their PI3K pathway status measured in arch... -

Phosphatidylinositol pathway
This cell signaling lecture explains about Phosphatidylinositol signal pathway. http://shomusbiology.com/ Download the study materials here- http://shomusbiology.com/bio-materials.html Remember Shomu’s Biology is created to spread the knowledge of life science and biology by sharing all this free biology lectures video and animation presented by Suman Bhattacharjee in YouTube. All these tutorials are brought to you for free. Please subscribe to our channel so that we can grow together. You can check for any of the following services from Shomu’s Biology- Buy Shomu’s Biology lecture DVD set- www.shomusbiology.com/dvd-store Shomu’s Biology assignment services – www.shomusbiology.com/assignment -help Join Online coaching for CSIR NET exam – www.shomusbiology.com/net-coaching We are social. F...
Dr. Hurvitz on Patient Outcomes and PIK3CA Mutations
- Order: Reorder
- Duration: 1:59
- Updated: 18 Jul 2014
- views: 80
- published: 18 Jul 2014
- views: 80
PIK3CA Mutational Status and Survival in Locally Advanced Cervical Cancer Patients
- Order: Reorder
- Duration: 8:34
- Updated: 16 Jul 2012
- views: 256
- published: 16 Jul 2012
- views: 256
Domain-specific PIK3CA mutations affect different pathway activities... - Christopher Benz
- Order: Reorder
- Duration: 15:01
- Updated: 23 May 2014
- views: 339
- published: 23 May 2014
- views: 339
PIK3CA Mutations in HER2-Positive Breast Cancer
- Order: Reorder
- Duration: 4:20
- Updated: 10 Feb 2014
- views: 481
- published: 10 Feb 2014
- views: 481
FISH analysis of HSR type PIK3CA amplification in endometrial cancer
- Order: Reorder
- Duration: 4:19
- Updated: 11 Dec 2014
- views: 163
- published: 11 Dec 2014
- views: 163
Randox KRAS BRAF PIK3CA* Array - Molecular Testing
- Order: Reorder
- Duration: 1:50
- Updated: 23 Dec 2014
- views: 191
- published: 23 Dec 2014
- views: 191
Are the benefits of aspirin in colorectal cancer limited to PIK3CA mutated cancers?
- Order: Reorder
- Duration: 9:28
- Updated: 21 Apr 2015
- views: 206
- published: 21 Apr 2015
- views: 206
Dr. Baselga Discusses PIK3CA Inhibitors in Breast Cancer
- Order: Reorder
- Duration: 1:02
- Updated: 28 Mar 2013
- views: 125
- published: 28 Mar 2013
- views: 125
The PI3K/AKT signalling pathway
- Order: Reorder
- Duration: 4:44
- Updated: 22 Feb 2010
- views: 205208
- published: 22 Feb 2010
- views: 205208
Role of TGF-beta signaling in PIK3CA-driven Head and Neck Cancer Invasion and Metastasis
- Order: Reorder
- Duration: 9:18
- Updated: 12 Sep 2013
- views: 337
- published: 12 Sep 2013
- views: 337
Sibylle Loibl: PIK3CA mutation predicts resistance to anti HER2/chemotherapy
- Order: Reorder
- Duration: 10:59
- Updated: 12 Dec 2013
- views: 138
- published: 12 Dec 2013
- views: 138
FISH analysis of a typical PIK3CA amplification in endometrial cancer.
- Order: Reorder
- Duration: 3:48
- Updated: 10 Dec 2014
- views: 96
- published: 10 Dec 2014
- views: 96
Aspirin Use, Tumor PIK3CA Mutation, and Colorectal Cancer Su
- Order: Reorder
- Duration: 3:08
- Updated: 21 Dec 2014
- views: 16
- published: 21 Dec 2014
- views: 16
Whitney Henry | Harvard Horizons Symposium
- Order: Reorder
- Duration: 6:05
- Updated: 21 May 2014
- views: 3458
- published: 21 May 2014
- views: 3458
Mario Campone
- Order: Reorder
- Duration: 6:22
- Updated: 11 Dec 2015
- views: 89
- published: 11 Dec 2015
- views: 89
Rapid Diagnostic Testing in Molecular Pathology
- Order: Reorder
- Duration: 4:21
- Updated: 24 Dec 2014
- views: 149
- published: 24 Dec 2014
- views: 149
Liquid Biopsy Clinical Research – Tracking Mutations in cfDNA and CTCs
- Order: Reorder
- Duration: 6:08
- Updated: 29 Feb 2016
- views: 408
- published: 29 Feb 2016
- views: 408
AACR 2009: Genetic influences in response to EGFR for metastatic colorectal cancer
- Order: Reorder
- Duration: 4:16
- Updated: 28 Oct 2009
- views: 715
- published: 28 Oct 2009
- views: 715
Dr. Kris Discusses Available Targeted Therapies
- Order: Reorder
- Duration: 0:31
- Updated: 10 Aug 2011
- views: 116
- published: 10 Aug 2011
- views: 116
Rare Mutation Detection w/ Digital PCR - ASHG 2014
- Order: Reorder
- Duration: 4:33
- Updated: 22 Oct 2014
- views: 1327
- published: 22 Oct 2014
- views: 1327
5 Women Who You Won’t Believe Actually Exist
- Order: Reorder
- Duration: 2:01
- Updated: 08 Mar 2016
- views: 44
- published: 08 Mar 2016
- views: 44
Buparlisib helps overcome endocrine resistance in metastatic breast cancer: BELLE-2 trial results
- Order: Reorder
- Duration: 7:02
- Updated: 06 Jan 2016
- views: 37
- published: 06 Jan 2016
- views: 37
Phosphatidylinositol pathway
- Order: Reorder
- Duration: 5:42
- Updated: 07 Dec 2012
- views: 8182
- published: 07 Dec 2012
- views: 8182
- Playlist
- Chat
- Playlist
- Chat

Dr. Hurvitz on Patient Outcomes and PIK3CA Mutations
- Report rights infringement
- published: 18 Jul 2014
- views: 80

PIK3CA Mutational Status and Survival in Locally Advanced Cervical Cancer Patients
- Report rights infringement
- published: 16 Jul 2012
- views: 256

Domain-specific PIK3CA mutations affect different pathway activities... - Christopher Benz
- Report rights infringement
- published: 23 May 2014
- views: 339

PIK3CA Mutations in HER2-Positive Breast Cancer
- Report rights infringement
- published: 10 Feb 2014
- views: 481

FISH analysis of HSR type PIK3CA amplification in endometrial cancer
- Report rights infringement
- published: 11 Dec 2014
- views: 163

Randox KRAS BRAF PIK3CA* Array - Molecular Testing
- Report rights infringement
- published: 23 Dec 2014
- views: 191

Are the benefits of aspirin in colorectal cancer limited to PIK3CA mutated cancers?
- Report rights infringement
- published: 21 Apr 2015
- views: 206

Dr. Baselga Discusses PIK3CA Inhibitors in Breast Cancer
- Report rights infringement
- published: 28 Mar 2013
- views: 125

The PI3K/AKT signalling pathway
- Report rights infringement
- published: 22 Feb 2010
- views: 205208

Role of TGF-beta signaling in PIK3CA-driven Head and Neck Cancer Invasion and Metastasis
- Report rights infringement
- published: 12 Sep 2013
- views: 337

Sibylle Loibl: PIK3CA mutation predicts resistance to anti HER2/chemotherapy
- Report rights infringement
- published: 12 Dec 2013
- views: 138

FISH analysis of a typical PIK3CA amplification in endometrial cancer.
- Report rights infringement
- published: 10 Dec 2014
- views: 96

Aspirin Use, Tumor PIK3CA Mutation, and Colorectal Cancer Su
- Report rights infringement
- published: 21 Dec 2014
- views: 16

Whitney Henry | Harvard Horizons Symposium
- Report rights infringement
- published: 21 May 2014
- views: 3458
-
Lyrics list:text lyricsplay full screenplay karaoke
Explosions Heard In Schaerbeek As New Anti-Terror Raid Launched In Brussels
Edit WorldNews.com 25 Mar 2016“Galvanized Girl” Is No “Comeback Kid”
Edit WorldNews.com 25 Mar 2016March Polls Show Sanders Would Defeat All Challengers If November Election Held Today
Edit WorldNews.com 25 Mar 2016Pope washes feet of Muslim migrants, says ‘We are brothers’
Edit The Hindu 25 Mar 2016Frontier Pharma: Innovative Licensing Opportunities in Non-Hodgkin Lymphoma
Edit PR Newswire 10 Mar 2016Transgenomic Launches New MX-ICP Liquid Biopsy Tests for Comprehensive Detection of Colorectal and Melanoma Tumor Mutations (Transgenomic Inc)
Edit Public Technologies 10 Mar 2016Study maps distinct molecular signatures of HPV-positive throat cancer patients by smoking status (ASTRO - ...
Edit Public Technologies 18 Feb 2016Transgenomic Launches New MX-ICP Panels for Liquid Biopsy Detection of RAS and PIK3CA Tumor Mutations ...
Edit Public Technologies 19 Jan 2016Memorial Sloan Kettering Studies Highlight Potential of Liquid Biopsy at San Antonio Breast Cancer Symposium ...
Edit Public Technologies 11 Dec 2015Yale study probes genes for clues to drug resistance in aggressive breast cancer (Yale University) ...
Edit Public Technologies 10 Dec 2015Caris Life Sciences Research Yields New Molecular Insights into Rare Breast Cancers Comprehensive Molecular Profiling Reveals Potentially Targetable Biomarkers in Metaplastic Breast Carcinoma and Malignant Phyllodes Tumors of the BreastSABCS Presentations Provide Further Evidence of Utility of Caris Molecular Intelligence in Characterizing Rare Breast Tumors
Edit PR Newswire 09 Dec 2015Novartis presents new data on targeted combination therapy at SABCS reinforcing commitment to breast cancer ...
Edit Public Technologies 02 Dec 2015Frontier Pharma Report 2015: Innovative Licensing Opportunities in Non-Hodgkin Lymphoma
Edit PR Newswire 12 Nov 2015Horizon Discovery introduces Cell Free DNA HDx™ Reference Standards (Horizon Discovery Group plc)
Edit Public Technologies 04 Nov 2015Moffitt Cancer Center Recognizes National Lung Cancer Awareness Month (H Lee Moffitt Cancer Center and ...
Edit Public Technologies 02 Nov 2015HER2 missense mutations have distinct effects on oncogenic signaling and migration
Edit PNAS 27 Oct 2015ArQule Presents Results From Phase 1b Expansion Study of ARQ 092 at the 2015 European ...
Edit Stockhouse 28 Sep 2015- 1
- 2
- 3
- 4
- 5
- Next page »







