- Order:
- Duration: 5:44
- Published: 07 Nov 2007
- Uploaded: 01 Sep 2011
- Author: dltv































![Progressive Field, home of the Cleveland Indians.The tradition of professional hockey in Cleveland started with the original Cleveland Barons in 1937.[99] Cleveland fielded an NHL team, also called the Cleveland Barons, from 1976 to 1978, which was later merged into the Minnesota North Stars (now the Dallas Stars). Progressive Field, home of the Cleveland Indians.The tradition of professional hockey in Cleveland started with the original Cleveland Barons in 1937.[99] Cleveland fielded an NHL team, also called the Cleveland Barons, from 1976 to 1978, which was later merged into the Minnesota North Stars (now the Dallas Stars).](http://web.archive.org./web/20110903061824im_/http://cdn4.wn.com/pd/50/3d/65a9b5ec557126cbe60998db9769_grande.jpg)

























| Name | Folding@home |
|---|---|
| Screenshot | |
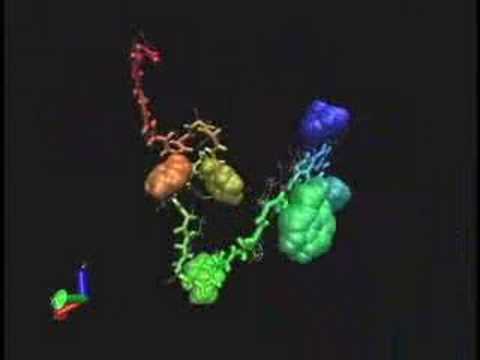
| Caption | The PlayStation 3 Folding@home client displays a 3D model of the protein being simulated. |
| Author | Vijay Pande |
| Developer | Stanford University / Pande Group |
| Released | 2000-10-01 |
| Latest release version | Windows:6.23 (uniprocessor)6.32 (GPU)Mac OS X:6.29.3 (PPC uniprocessor)6.29.3 (x86 SMP)Linux:6.02 (uniprocessor)6.34 (x86-64 SMP)PlayStation 3: 1.3.1 |
| Latest release date | (Mac SMP) |
| Latest preview version | All platforms:7.1.24 |
| Latest preview date | |
| Platform | Cross-platform |
| Language | English |
| Genre | Distributed computing |
| License | Proprietary |
| Website |
Folding@home ("Folding at Home", FAH, F@h) is a distributed computing (DC) project designed to perform computationally intensive simulations of protein folding and other molecular dynamics (MD), and to improve on the methods available to do so. It was launched on October 1, 2000, and is currently managed by the Pande Group, within Stanford University's chemistry department, under the supervision of Professor Vijay Pande.
In 2007 Guinness recognized Folding@home as the most powerful distributed computing cluster in the world. However, Bitcoin is now thought to be the most powerful distributed computer cluster. Folding@home is one of the world's largest distributed computing projects. The goal of the project is "to understand protein folding, misfolding, and related diseases."
Accurate simulations of protein folding and misfolding enable the scientific community to better understand the development of many diseases, including sickle-cell disease (drepanocytosis), Alzheimer's disease, Parkinson's disease, Bovine spongiform encephalopathy, cancer, Huntington's disease, cystic fibrosis, osteogenesis imperfecta, alpha 1-antitrypsin deficiency, and other aggregation-related diseases. More fundamentally, understanding the process of protein folding — how biological molecules assemble themselves into a functional state — is one of the outstanding problems of molecular biology. So far, the Folding@home project has successfully simulated folding in the 1.5 millisecond range — which is a simulation thousands of times longer than it was previously thought possible to model.
The Pande Group's goal is to refine and improve the MD and Folding@home DC methods to the level where it will become an essential tool for MD research, and to achieve that goal they collaborate with various scientific institutions. As of November 23, 2010, seventy-eight scientific research papers have been published using the project's work. A University of Illinois at Urbana-Champaign report dated October 22, 2002 states that Folding@home distributed simulations of protein folding are demonstrably accurate.
The Folding@home client periodically connects to a server to retrieve "work units", which are packets of data upon which to perform calculations. Each completed work unit is then sent back to the server. As data integrity is a major concern for all distributed computing projects, all work units are validated through the use of a 2048 bit digital signature.
Contributors to Folding@home may have user names used to keep track of their contributions. Each user may be running the client on one or more CPUs; for example, a user with two computers could run the client on both of them. Users may also contribute under one or more team names; many different users may join together to form a team. Contributors are assigned a score indicating the number and difficulty of completed work units. Rankings and other statistics are posted to the Folding@home website http://folding.stanford.edu/.
Interest and participation in the project has grown steadily since its launch. The number of active devices participating in the project increased substantially after receiving much publicity during the launch of their high performance clients for both ATi graphics cards and the PlayStation 3, and again following the launch of the high performance client for nVidia graphics cards.
As of March 28, 2011 the peak speed of the project overall has reached over 7 native PFLOPS (12.4 x86 PFLOPS) from around 464,000 active machines, and the project has received computing results from over 6.07 million devices since it first started.
Starting in April 2009, Folding@home began reporting performance in both "Native" FLOPS and x86 FLOPS. which uses AMD/ATI's CAL. On June 17, 2008, a version of the second-generation Windows GPU client for CUDA enabled Nvidia GPUs was also released for public beta testing. The GPU clients proved reliable enough to be promoted out of the beta phase and were officially released August 1, 2008. Newer GPU cores continue to be released for both CAL and CUDA.
While the only officially released GPU v2.0 client is for Windows, this client can be run on Linux under Wine with NVIDIA graphics cards. The client can operate on both 32- and 64-bit Linux platforms, but in either case the 32-bit CUDA toolkit is needed. This configuration is not officially supported, though initial results have shown comparable performance to that of the native client and no problems with the scientific results have been found. An unofficial installation guide has been published. GPU3 will use OpenCL (preferred over DirectCompute) as the software interface, which may mean that the GPU core will be unified for both ATI and nVidia, and may also mean the addition of support for other platforms with OpenCL support.
On May 25, 2010, Vijay Pande announced an open beta of the GPU3 client on the Folding@home blog. The new core initially supports only Nvidia GPUs, but will support ATI/AMD GPUs in a subsequent release.
On November 13, 2006, the beta SMP Folding@home clients for x86-64 Linux and x86 Mac OS X were released. The beta win32 SMP Folding@home client is out as well, and a 32-bit Linux client is currently in development.
On June 17, 2009 the Pande Group revealed that a second generation SMP client (known as the SMP2 client) was in development. This client will use threads rather than MPI Some teams offer prizes in an attempt to increase participation in the project.
A development version of Folding@home once ran on the open source BOINC framework; however, this version remained unreleased.
The total power needed to produce the processing power used by the project can be estimated based on the average FLOPS per watt. As of November 2008, according to the Green500 list, the most efficient computer - also based on a version of the Cell BE - runs at 536.24 MFLOPS/watt. One petaFLOPS equals 1,000,000,000 MFLOPSs. Therefore, the current Folding@home project, if it were theoretically using the most efficient CPUs currently available, would use at least 2.8 megawatts of power per petaFLOPS, slightly more than the world's first petaFLOPS system, the Cell-based Roadrunner which uses 2.345MW. This is equivalent to the power needed to light approximately 40,000 standard house light bulbs (between 60 and 100 watts each), or the equivalent of 1-3 wind turbines depending on their size.
Estimates of energy usage per time period are more difficult than estimates of energy usage per processing instruction. This is because Folding@home clients are often run on computers that would be powered-on even in the absence of the Folding@home client, and that run other programs simultaneously.
Looking at energy-balance in a larger context needs a number of assumptions. While Folding@home increases processor utilization, and thus (usually) power use; the extent to which it does so depends on the client processor's normal operating load, and its ability to reduce clock speeds when presented with less-than-full utilization (a process known as dynamic frequency scaling). In a Folding@home client that runs in a heated home, the excess heat generated by the power usage would reduce the amount of energy needed to heat the building somewhat. However, such energy gains would be offset over the year in those locations where air-conditioners are used during warmer months.
Category:Protein folds Category:Protein structure Category:Distributed computing projects Category:Windows software Category:Linux science software Category:Mac OS X software Category:PlayStation 3 software Category:Cross-platform software Category:2000 introductions
This text is licensed under the Creative Commons CC-BY-SA License. This text was originally published on Wikipedia and was developed by the Wikipedia community.



